ABSTRACT
Research reproducibility is essential for scientific development. Yet, rates of reproducibility are low. As increasingly more research relies on computers and software, efforts for improving reproducibility rates have focused on making research products digitally available, such as publishing analysis workflows as computer code, and raw and processed data in computer readable form. However, research products that are digitally available are not necessarily friendly for learners and interested parties with little to no experience in the field. This renders research products unapproachable, counteracts their availability, and hinders scientific reproducibility. To improve both short- and long-term adoption of reproducible scientific practices, research products need to be made approachable for learners, the researchers of the future. Using a case study within evolutionary biology, we identify aspects of research workflows that make them unapproachable to the general audience: use of highly specialized language; unclear goals and high cognitive load; and lack of trouble-shooting examples. We propose principles to improve the unapproachable aspects of research workflows and illustrate their application using an online teaching resource. We elaborate on the general application of these principles for documenting research products and teaching materials, to provide present learners and future researchers with tools for successful scientific reproducibility. Supplementary materials for this article are available online.
1 Introduction
Research reproducibility—the extent to which consistent results are obtained when a scientific experiment or research workflow is repeated (Curating for Reproducibility Consortium Citation2017)—is a key aspect of the advancement of science, as it constitutes a minimum standard that allows understanding research products, that is, methods, data, analysis, results, etc. (Piwowar Citation2013), to determine their reliability and generality, and eventually build up scientific knowledge and applications based on those products (King Citation1995; Peng Citation2011; Powers and Hampton Citation2019). In the natural sciences, rates of reproducibility are low (Ioannidis Citation2005; Prinz, Schlange, and Asadullah Citation2011), which has elicited concerns about a crisis in the field (Baker Citation2016).
In response, the scientific community has been developing new principles and standards to incentivize cultural changes that support a long-term improvement of reproducibility rates in the natural sciences (Peng Citation2015; Wilkinson et al. Citation2016; Miyakawa Citation2020). A standard for reproducibility that has received much attention is availability, which we define as a property denoting that a research product can be reached (acquired, copied, analyzed, processed and/or reused) at no financial, legal or technical cost (The Turing Way Community Citation2021), and without geographic, demographic, social or temporal barriers for the population (Fecher and Friesike Citation2014).
In this article, we argue that research products that are digitally available are often unapproachable in practice, because they are not friendly for learners and interested parties with different levels of experience in the field. Research products that are unapproachable counteract availability, and hinder reproducibility short and long term. To support long-term adoption of reproducible practices in the natural sciences, research workflows need to be made approachable for learners, the researchers of the future (Roland et al. Citation2002; National Academies of Sciences, Engineering, and Medicine Citation2018).
To elaborate on our thesis, we designed a case study within the research field of phylogenetics, a discipline within evolutionary biology. We use our case study to identify barriers that have made research workflows largely unapproachable to a general audience in the natural sciences. Then, we propose some principles for researchers to address these barriers and create research workflows that are reproducible by a larger audience. The principles proposed here can be generalized and integrated into the undergraduate and graduate school STEM curriculum, either for courses specialized in reproducibility or within other subject areas, as a necessary component of successful and impactful science.
2 A Case Study from Phylogenetics
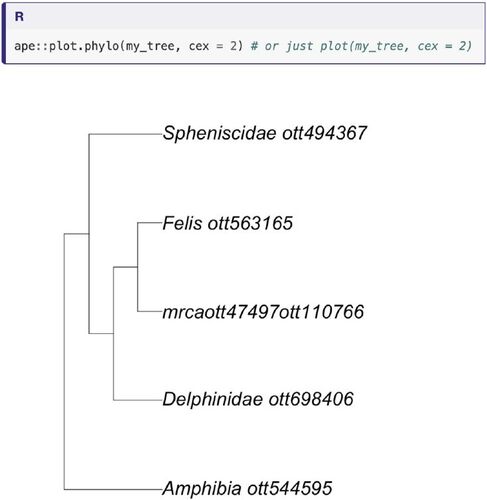
Phylogenetics is a key discipline within evolutionary biology (Dobzhansky Citation1973). It focuses on investigating the history of shared ancestry of living and extinct organisms using biological data and represents this evolutionary history with a diagram known as a phylogeny or phylogenetic tree (because it grows through time and appears to have branches; ). Phylogenies provide the basis to study and understand all biological processes in an evolutionary context (Dobzhansky Citation1973). Hence, it follows that improving reproducibility rates in phylogenetics has the potential to positively impact research across the natural sciences.
Fig. 1 A phylogenetic tree from our tutorial. It was extracted using OpenTree of Life resources (Open Tree Of Life et al. Citation2019) wrapped in the rotl R package (Michonneau, Brown, and Winter Citation2016).
To explore barriers to approachable phylogenetics, we develop a case study that touches on three common problems within the field: standardizing organism names in phylogenies, obtaining current phylogenetic knowledge for a group of organisms, and summarizing this phylogenetic knowledge in a meaningful way. To address these problems, we propose a research workflow that relies on resources from the Open Tree of Life (OpenTree), an open source project that provides digital availability of phylogenetic results from published, peer-reviewed research, which is considered to be vetted and state-of-the-art knowledge in the field. OpenTree phylogenies are stored in a public database, the Phylesystem (McTavish et al. Citation2015), and are downloadable as various computer-readable file types, which is key for reusable and reproducible workflows (Wilson et al. Citation2017). OpenTree also provides access to a single standard for organism names (taxonomic standard) that is applied to the stored phylogenies (Rees and Cranston Citation2017), which are then used to summarize a single phylogenetic tree encompassing all life (Open Tree Of Life et al. 2019).
All of these resources are available for download and use from OpenTree, free of financial cost to any user. One way to access OpenTree resources is manually, through its Graphical User Interface (GUI; aka, a website or application that allows users to access and use functionalities with mouse or keyboard clicks). However, reducing as many manual steps as possible in research workflows is key for reproducibility, as manual data manipulation scales poorly and is prone to error (Bakken Citation2019). OpenTree’s resources are also programmatically available through its Application Programming Interface services (APIs; aka, computer code that automatically implements functionalities, that is usually used by programmers to build different or tailored functionalities). While APIs provide data processing scalability and reproducibility (Open Tree Of Life et al. Citation2016), they come at a high technical and cognitive cost for the user, whom requires considerably more computer programming experience and literacy to be able to successfully use APIs. OpenTree’s API services have been wrapped by the rotl R package (Michonneau, Brown, and Winter Citation2016) and the opentree Python module (McTavish, Sánchez Reyes, and Holder Citation2021). R and Python programming languages are open source and free of cost and represent two of the most widely used programming languages in the sciences today (Eglen Citation2009; Baker Citation2017). As such, rotl and opentree software packages are contributing to approachability of OpenTree’s resources to R and Python users, increasing availability to a wider user base.
However, while learners in the natural sciences have been engaging independently with R and Python programming languages, computer programming is not traditionally a core skill formally taught to biologists and naturalists (Sayres et al. Citation2018; Wright et al. Citation2019; Williams et al. Citation2019). As computers continue to play a larger role in most scientific disciplines (Piccolo and Frampton Citation2016), higher baseline computational skills are required across all natural sciences not only to develop an original research workflow, but to be able to follow and reproduce research workflows from other researchers (National Academies of Sciences, Engineering, and Medicine Citation2019).
Thus, efforts to increase reproducibility rates long term in the natural sciences would benefit from addressing specific barriers for learners in the field, to support them in acquiring the skills needed to reproduce research workflows that rely heavily on computer code (Peng Citation2011; Sandve et al. Citation2013; Powers and Hampton Citation2019).
In the next section, we describe (in no particular order) three barriers to approachable research workflows that we identify using our case study. Then we develop a set of principles to address these barriers and apply the latter to a set of teaching materials that are available at https://mctavishlab.github.io/R_OpenTree_tutorials/.
3 Identifying Barriers to Approachable Research Workflows
The main goal of our case study is to obtain a single phylogeny summarizing data from a set of published phylogenies for the canids (the family of dogs, coyotes, wolves, etc.), our organisms of study. All analysis for our case study can be completely accomplished using functions from the R package rotl or the Python module opentree. If a researcher were to use the proposed analysis workflow in a publication, they would typically describe it in the methodology section as “The canid summary phylogeny was obtained using functions from X package, details are available as supplementary materials.” This is usual practice, mainly because journals do not have space to publish all code used for an analysis in the methods section. Yet, supplementary materials and data have the misfortune to not be peer-reviewed as thoroughly (or at all) as the main manuscript (Pop and Salzberg Citation2015). They are also prone to the dreaded promise “available upon request,” which has very low rates of fulfillment (Krawczyk and Reuben Citation2012; Gabelica, Bojčić, and Puljak Citation2022). Without the primary data and code that was used to perform an analysis, it is impossible to reproduce said analysis (Miyakawa Citation2020). When the code is available, other issues can complicate reproduction of the analysis, to the point of completely obstructing reproducibility. For example, some questions that are often unaddressed in research workflows are: What software do I need to read the code files? What software can I use to actually run the code? Do I need additional software that the analysis depends on? What software versions were used? What does the code even mean?
Some of these questions can be answered by referring to the software documentation, which is usually publicly available and can be accessed by any potential user. As opposed to code, software documentation is written in natural language (i.e., any known human language, e.g., English, Spanish, Chinese) and is considered a key element for successful adoption of software by target users (Karimzadeh and Hoffman Citation2018). This might explain why documentation for software addressed to academic users is also usually written using highly specialized computational language or jargon (i.e., computationally specific concepts, words, and phrases) as well as formal scientific and academic language. We identify this as barrier 1 to approachable research workflows—Specialized language is intimidating. While scientific jargon might have an important role for formal acceptance of software by the scientific and academic community, it can be perceived as cold and/or intimidating language that often slows down or even obstructs examination, application, and adoption of code by a wider audience (Ball Citation2017). In contrast, introducing information without the use of jargon supports learner’s conceptual understanding of new ideas and concepts (McDonnell, Barker, and Wieman Citation2016; Pan et al. Citation2019).
Another element of good software documentation is that it has to be thorough (Karimzadeh and Hoffman Citation2018), meaning that it should describe general usage of individual functions, as well as arguments and variables that said function can take (Karimzadeh and Hoffman Citation2018). Individual documentation for each function is usually presented in alphabetic order and does not have a specific analysis goal. Moreover, most software has numerous functions, so documentation is usually very lengthy and it is hard to navigate. This can have the effect of increasing the amount of information that needs to be simultaneously processed by the users, which can lead to overload of the finite amount of working memory any one possesses, known as cognitive load (Sweller Citation1988). In this context, identifying connections across functions that are meant to work on the same analysis workflow can become a very difficult task. We recognize this as barrier 2—Lack of specific goals leading to high cognitive load. High cognitive load is know to have a negative effect in learning software (Chandler and Sweller Citation1996; Van Merriënboer and Ayres Citation2005; Lambert, Kalyuga, and Capan Citation2009).
A third important aspect of software documentation are examples that demonstrate usage of individual functions (Karimzadeh and Hoffman Citation2018). Examples presented in software documentation are usually worked to perfection, as they are intended to showcase the ideal or minimal case in which a function works well. Perfectly worked examples ignore the user experience by maintaining focus on the software content and fail to provide users with expert and clear advice on how to troubleshoot if needed. We identify this as barrier 3—Lack of trouble-shooting examples. Error management training is an approach that focuses on framing mistakes as beneficial to learning complex tasks, to give learners the opportunity to actively explore a task with a positive mindset (Frese Citation1995). Providing examples that showcase potential errors, supports user’s performance (Steele-Johnson and Kalinoski Citation2014), and can greatly improve learner’s ability to troubleshoot outside the classroom (Shannon and Summet Citation2015; Nederbragt et al. Citation2020).
In sum, best practices for good software documentation are not enough to promote reproducibility of published research workflows that rely heavily on code. In the following section, we describe some principles that can help to reduce or remove the identified barriers, to create research workflows that are more approachable and hence more reproducible by a larger audience.
4 Principles for Approachable Research Workflows
4.1 Principle 1. Use Friendly, Relatable and Respectful Language
Avoiding formal language, and incorporating elements of pop culture, such as picture character icons known as “emojis,” make the language more familiar to a broader target audience (). We made an effort to specifically complement the primary documentation by identifying computational concepts that were assumed or were not explained in depth. We vetted the tutorials through feedback from workshop participants as well as individual users to identify such specialized concepts.
4.2 Principle 2. Reduce Cognitive Load by Providing Specific and Clear Goals with Literate Programming
Cognitive load can be greatly reduced for learners by applying an active learning strategy such as linking usage to a “real world” or “human” application (Felder and Brent Citation2009). Programming computer languages are by themselves quite abstract and represent a learning subject with a potentially high cognitive load for most learners. Pedagogical research shows that active learning practices are one of the most effective ways to take on abstract subjects (Freeman et al. Citation2014). A story-like narrative that links code usage in an integrative example, invites learners to try the code, which can lead them to remember what they are doing and why they are doing it. This “literate programming” paradigm (Knuth Citation1984; Fritzson, Gunnarsson, and Jirstrand Citation2002) makes code more approachable, as it integrates narratives with computer code in the same document, supporting learners in actively following the code usage, supporting memory and understanding (Piccolo and Frampton Citation2016).
We propose that documents developed with “literate programming” can be made more accessible by choosing narratives that are relatable to a more general audience. An easy way to do this in biology is choosing a charismatic taxon as a model organism. For a research group, this can be the biological group they are studying. For the general audience, a highly charismatic group—such as dinosaurs, should work well. For example, when we presented our tutorial to the Amphibia Web Organization (van der Meijden et al. Citation2002) in January 2020, we tailored all examples to frogs and their allies.
We examined available software documentation for the package rotl and designed a narrative that requires the usage of as many functions as possible. We demonstrate code applications that are commonly requested by OpenTree users, but that are not demonstrated in the documentation of the package. By framing the function workflow using highly requested uses, the documentation acquires a narrative arc that is easier to follow and remember by users. This can also facilitate translating the code application to other use cases of interest for learners in biology.
4.3 Principle 3. Provide Examples That Are User-focused by Demonstrating Errors and Warnings
An activity that has become increasingly widespread in programming-language education is live programming. During live programming, an instructor writes code and executes it in a way that is visible to learners through a screen (Guzdial and Barr Citation2013; Selvaraj et al. Citation2021). One benefit of this practice is that typos and mistakes occur, normalizing them for learners. Watching an instructor handling errors, demonstrates learners on how to solve them when they are outside the classroom (Shannon and Summet Citation2015; Nederbragt et al. Citation2020). When coauthor McTavish was a postdoc, teaching an introductory programming workshop as a volunteer with the Carpentries (a nonprofit group that teaches foundational coding and data science skills to researchers worldwide; Wilson Citation2006, Citation2022), a senior faculty member taking the workshop complained that the typos were slowing things down and interfering with the pedagogy. McTavish replied “the typos ARE the pedagogy.” This has become a slogan of sorts at the Carpentries, capturing the idea that embracing and discussing mistakes is essential to teaching programming (Wilson Citation2019). Yet, working through mistakes is rarely done on written pedagogical materials. Software documentation focuses on demonstrating usage function with examples that work seamlessly, without errors. We argue that the opposite is needed to support adoption of reproducible workflows and support long term independence in learner’s and user’s performance (Gaspar and Langevin Citation2007; Steele-Johnson and Kalinoski Citation2014).
In our tutorials, we apply this principle by demonstrating examples that do not work as expected, and exemplifying ways to address issues (). For example, we identified inputs that would give a wide range of warnings and errors. We then focus on providing explanations for these errors and warning messages. We believe this supports users and learners to be less intimidated by the messages, and to practice taking useful information out of them.
We also demonstrate ways to evaluate inputs to determine if they will trigger an error or warning, and design and demonstrate alternative analysis routes on what to do when faced with an error or warning. One of the most essential skills in programming is interpreting and moving forward from errors. This has two pedagogical benefits. First, it provides users and learners with the means to troubleshoot their own warnings and errors. Second, it allows them to understand with more depth what the code is doing.
5 Conclusion
Response from the community has been invaluable in gauging success of our teaching materials. Senior researchers often comment on the usefulness of the tutorials for their research, as well as how they have supported students in using the demonstrated R packages more independently. We note that making approachable research workflows has all the advantages of reproducible research workflows. It saves time during explanation and training, when analyses are run by new collaborators and students. It can also save research time for yourself, when analyses are run again with more data, a different dataset, a different organism or biological model. It contributes to making scientific efforts that can build off of each other.
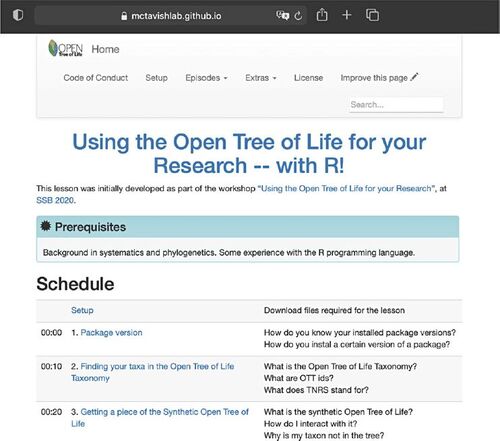
When developing our tutorials, we not only applied the principles elaborated here to make them more approachable, but we followed basic recommendations for successful reproducibility (Sandve et al. Citation2013). Applying all these principles in a tutorial not only teaches reproducibility, but also makes the teaching process itself reproducible (Dogucu and Cetinkaya-Rundel Citation2022). For example, we published the tutorials on a persistent and public website (Sánchez Reyes, McTavish, and Holder Citation2021) that adheres to the four r’s of openness, by being free license, free of cost, and thus free for use, reuse, redistribute, revise and remix (Hilton III et al. Citation2010). To make the website persistent, any updates to the tutorial are published as new versions. Versions presented at workshops are a copy from the original repository, and constitute a temporally stable snapshot of functions and workflows presented during a live workshop (Wilson Citation2006, Citation2022). This ensures that the tutorials are available for the users to return to any time they need it, and to be shared with other users and learners (). Finally, we dedicate a section of the tutorial to document the software versions that are demonstrated throughout. A common issue in open source software packages written in R and Python is the deprecation of functions (i.e., functions that are no longer reccommended or maintained, and that are in the process of being phased out and replaced by new ones; Marks et al. Citation2017; Vadlamani, Kalicheti, and Chimalakonda Citation2021). Running code that has fallen victim to deprecation require considerable programming savvy, and it is a task that is hard even for experienced programmers (Vadlamani, Kalicheti, and Chimalakonda Citation2021). Yet, old versions of open source software packages remain digitally accessible, and persist in time. If a package version is known, an analysis can be reproduced without investing additional resources in finding the appropriate functions to run it, even long time after a function had been deprecated.
Fig. 3 Snapshot of the home to our tutorial website, showing part of the schedule. Our tutorial website was constructed using the software workshop template from the Carpentries (Wilson Citation2016).
Incorporating the principles described here into teaching resources not only improves reproducibility practices, but it should facilitate adoption of software and analysis workflows in the natural sciences, among researchers at different academic levels, from undergrads to established researchers. It can also help close the academic gap that is generated by uneven access to computational resources across students belonging to different groups (KewalRamani et al. Citation2018), where the most affected learners usually belong to underrepresented minorities and rural areas (Warner et al. Citation2021). Differences in access can also be due to gender-biased parental and community pressures, in which male identifying individuals are more likely to be encouraged to perform activities related to computers (Google Inc. and Gallup Inc. Citation2016), while female identifying individuals are discouraged, starting from as early as elementray school (Master, Meltzoff, and Cheryan Citation2021).
Some universities and academic groups have started to incorporate reproducibility as a subject into their curriculum (NIGMS Career Curriculum Development Citation2015; Debruine and Taylor Citation2019; University of Washington Libraries Citation2022). The focus of these resources has been for students to acquire and practice skills to document their work. The principles identified and outlined here can be used to set learning goals and outcomes for new reproducibility syllabi. Ultimately, the long term improvement of reproducibility rates in science will depend on our ability to intentionally integrate best practices for achieving reproducibility into the educational framework of future data scientists (National Academies of Sciences, Engineering, and Medicine Citation2018). Inclusion of reproducibility into the data acumen of undergraduate curriculum will provide college learners and future researchers with the tools to develop the fundamental skills needed to successfully create reproducible scientific workflows and research products.
Acknowledgments
The authors gratefully acknowledge “Sustaining the Open Tree of Life”, NSF ABI No. 1759838, and ABI No. 1759846. They are also deeply grateful toward “The Carpentries” organization; without its invaluable volunteers and resources this project could not have been possible
Supplementary Materials
Title: Website and GitHub repository containing the complete teaching materials developed and demonstrated here.
GitHub repository link
Website link
Additional information
Funding
References
- Baker, M. (2016), “Is There a Reproducibility Crisis?,” Nature, 533, 353–366.
- Baker, M (2017), “Scientific Computing: Code Alert,” Nature, 541, 563–565.
- Bakken, S. (2019), “The Journey to Transparency, Reproducibility, and Replicability,” Journal of the American Medical Informatics Association, 26, 185–187. DOI: 10.1093/jamia/ocz007.
- Ball, P. (2017), “It’s Not Just You: Science Papers Are Getting Harder to Read,” Nature, 30. [online] Available at https://www.nature.com/news/it-s-not-just-you-science-papers-are-getting-harder-to-read-1.21751
- Chandler, P., and Sweller, J. (1996), “Cognitive Load While Learning to Use a Computer Program,” Applied Cognitive Psychology, 10, 151–170. DOI: 10.1002/(SICI)1099-0720(199604)10:2<151::AID-ACP380>3.0.CO;2-U.
- Curating for Reproducibility Consortium. (2017), “Defining ‘reproducibility’,” Available at cure.web.unc.edu/defining-reproducibility/.
- Debruine, L., and Taylor, J. (2019), “PsyTeachR - University of Glasgow School of Psychology and Neuroscience,” Available at https://psyteachr.github.io/.
- Dobzhansky, T. (1973), “Nothing in Biology Makes Sense Except in the Light of Evolution,” The American Biology Teacher, 35, 125–129. DOI: 10.2307/4444260.
- Dogucu, M., and Cetinkaya-Rundel, M. (2022), “Tools and Recommendations for Reproducible Teaching,” Technical Report. arXiv:2202.09504 [stat] type: article. Available at http://arxiv.org/abs/2202.09504.
- Eglen, S. J. (2009), “A Quick Guide to Teaching R Programming to Computational Biology Students,” PloS Computational Biology, 5, e1000482. DOI: 10.1371/journal.pcbi.1000482.
- Fecher, B., and Friesike, S. (2014), Open Science: One Term, Five Schools of Thought, pp. 17–47, Cham: Springer.
- Felder, R. M., and Brent, R. (2009), “Active Learning: An Introduction,” ASQ Higher Education Brief, 2, 1–5.
- Freeman, S., Eddy, S. L., McDonough, M., Smith, M. K., Okoroafor, N., Jordt, H., and Wenderoth, M. P. (2014), “Active Learning Increases Student Performance in Science, Engineering, and Mathematics,” Proceedings of the National Academy of Sciences, 111, 8410–8415. DOI: 10.1073/pnas.1319030111.
- Frese, M. (1995), “Error Management in Training: Conceptual and Empirical Results,” in Organizational Learning and Technological Change,” Springer, pp. 112–124.
- Fritzson, P., Gunnarsson, J., and Jirstrand, M. (2002), “MathModelica – An Extensible Modeling and Simulation Environment With Integrated Graphics and Literate Programming,” in 2nd International Modelica Conference, March 18-19, Munich, Germany.
- Gabelica, M., Bojčić, R., and Puljak, L. (2022), “Many Researchers Were Not Compliant With Their Published Data Sharing Statement: Mixed-Methods Study,” Journal of Clinical Epidemiology, 150, 33–41. DOI: 10.1016/j.jclinepi.2022.05.019.
- Gaspar, A., and Langevin, S. (2007), “Restoring “Coding With Intention” in Introductory Programming Courses,” in Proceedings of the 8th ACM SIGITE Conference on Information Technology Education, pp. 91–98.
- Google Inc., and Gallup Inc. (2016), “Diversity Gaps in Computer Science: Exploring the Underrepresentation of Girls, Blacks and Hispanics,” Retrieved from http://goo.gl/PG34aH (Additional reports from Google’s Computer Science Education Research are available at g.co/cseduresearch).
- Guzdial, M., and Barr, V. (2013), “The Lure of Live Coding; the Attraction of Small Data.” Communications of the ACM (Association of Computing Machinery), 56, 10–11. DOI: 10.1145/2534706.2534710.
- Hilton III, J., Wiley, D., Stein, J., and Johnson, A. (2010), “The Four “R’s of Openness and ALMS Analysis: Frameworks for Open Educational Resources,” Open Learning: The Journal of Open, Distance and e-Learning, 25, 37–44.
- Ioannidis, J. P. (2005), “Why Most Published Research Findings Are False,” PloS Medicine, 2, e124. DOI: 10.1371/journal.pmed.0020124.
- Karimzadeh, M., and Hoffman, M. M. (2018), “Top Considerations for Creating Bioinformatics Software Documentation,” Briefings in Bioinformatics,19, 693–699. DOI: 10.1093/bib/bbw134.
- KewalRamani, A., Zhang, J., Wang, X., Rathbun, A., Corcoran, L., Diliberti, M., and Zhang, J. (2018), “Student Access to Digital Learning Resources outside of the Classroom. NCES 2017-098,” National Center for Education Statistics.
- King, G. (1995), “Replication, Replication,” PS: Political Science & Politics, 28, 444–452.
- Knuth, D. E. (1984), “Literate Programming,” The Computer Journal, 27, 97–111. DOI: 10.1093/comjnl/27.2.97.
- Krawczyk, M., and Reuben, E. (2012), “(Un) Available Upon Request: Field Experiment on Researchers’ Willingness to Share Supplementary Materials,” Accountability in Research, 19, 175–186.
- Lambert, J., Kalyuga, S., and Capan, L. A. (2009), “Student Perceptions and Cognitive Load: What Can They Tell Us About e-Learning Web 2.0 Course Design?,” e-Learning and Digital Media, 6, 150–163. DOI: 10.2304/elea.2009.6.2.150.
- Marks, S., Buckley, A., Reinhold, M., and Goetz, B. (2017), “JEP 277: Enhanced Deprecation,” JEP 277: Enhanced Deprecation. Available at http://openjdk.java.net/jeps/277.
- Master, A., Meltzoff, A. N., and Cheryan, S. (2021), “Gender Stereotypes About Interests Start Early and Cause Gender Disparities in Computer Science and Engineering,” Proceedings of the National Academy of Sciences, 118, e2100030118. DOI: 10.1073/pnas.2100030118.
- McDonnell, L., Barker, M. K., and Wieman, C. (2016), “Concepts First, Jargon Second Improves Student Articulation of Understanding,” Biochemistry and Molecular Biology Education, 44, 12–19. DOI: 10.1002/bmb.20922.
- McTavish, E. J., Hinchliff, C. E., Allman, J. F., Brown, J. W., Cranston, K. A., Holder, M. T., Rees, J. A., and Smith, S. A. (2015), ‘Phylesystem: A Git-Based Data Store for Community-Curated Phylogenetic Estimates,” Bioinformatics, 31, 2794–2800. DOI: 10.1093/bioinformatics/btv276.
- McTavish, E. J., Sánchez Reyes, L. L., and Holder, M. T. (2021), “OpenTree: A Python Package for Accessing and Analyzing Data from the Open Tree of Life,” Systematic Biology, 70, 1295–1301. 10.1093/sysbio/syab033.
- Michonneau, F., Brown, J. W., and Winter, D. J. (2016), “rotl: an R Package to Interact With the Open Tree of Life Data,” Methods in Ecology and Evolution, 7, 1476–1481. DOI: 10.1111/2041-210X.12593.
- Miyakawa, T. (2020), “No Raw Data, No Science: Another Possible Source of the Reproducibility Crisis.” Molecular Brain, 13, 1–6. DOI: 10.1186/s13041-020-0552-2.
- National Academies of Sciences, Engineering, and Medicine. (2018), Data Science for Undergraduates: Opportunities and Options, Washington, DC: National Academies Press.
- National Academies of Sciences, Engineering, and Medicine (2019), Reproducibility and Replicability in Science, Washington, DC: National Academies Press.
- Nederbragt, A., Harris, R. M., Hill, A. P., and Wilson, G. (2020), “Ten Quick Tips for Teaching With Participatory Live Coding,” PloS Computational Biology, 16, e1008090. DOI: 10.1371/journal.pcbi.1008090.
- NIGMS Career Curriculum Development. (2015), “Rigor & Reproducibility, National Institute of General Medical Sciences.” Available at https://www.nigms.nih.gov/training/instpredoc/Pages/admin-supplements-prev.aspx.
- Open Tree Of Life, Redelings, B., Cranston, K. A., Allman, J., Holder, M. T., and McTavish, E. J. (2016), “Open Tree of Life APIs v3.0,” Open Tree of Life Project (online resources). Available at https://github.com/OpenTreeOfLife/germinator/wiki/Open-Tree-of-Life-Web-APIs.
- Open Tree Of Life, Redelings, B., Sánchez Reyes, L. L., Cranston, K. A., Allman, J., Holder, M. T., and McTavish, E. J. (2019), “Open Tree of Life Synthetic Tree v12.3,” Zenodo. Available at DOI: 10.5281/zenodo.3937742..
- Pan, S. C., Cooke, J., Little, J. L., McDaniel, M. A., Foster, E. R., Connor, L. T., and Rickard, T. C. (2019), “Online and Clicker Quizzing on Jargon Terms Enhances Definition-Focused but not Conceptually Focused Biology Exam Performance,” CBE-Life Sciences Education, 18, ar54. DOI: 10.1187/cbe.18-12-0248.
- Peng, R. (2015), “The Reproducibility Crisis in Science: A Statistical Counterattack,” Significance, 12, 30–32. DOI: 10.1111/j.1740-9713.2015.00827.x.
- Peng, R. D. (2011), “Reproducible Research in Computational Science,” Science, 334, 1226–1227. DOI: 10.1126/science.1213847.
- Piccolo, S. R., and Frampton, M. B. (2016), “Tools and Techniques for Computational Reproducibility,” Gigascience, 5, S13742–S016. DOI: 10.1186/s13742-016-0135-4.
- Piwowar, H. (2013), “Value All Research Products,”’ Nature, 493, 159–159. DOI: 10.1038/493159a.
- Pop, M., and Salzberg, S. L. (2015), “Use and Mis-Use of Supplementary Material in Science Publications.” BMC Bioinformatics, 16, 1–4.
- Powers, S. M., and Hampton, S. E. (2019), “Open Science, Reproducibility, and Transparency in Ecology,” Ecological Applications, 29, e01822.
- Prinz, F., Schlange, T., and Asadullah, K. (2011), “Believe it or Not: How Much Can We Rely on Published Data on Potential Drug Targets?,” Nature Reviews Drug Discovery, 10, 712–712.
- Rees, J. A., and Cranston, K. (2017), “Automated Assembly of a Reference Taxonomy for Phylogenetic Data Synthesis,” Biodiversity Data Journal 5, e12581.
- Roland, M.-C., Chèvre, A.-M., Chadoeuf, J., Hubert, B., and Bonnemaire, J. (2002), “Think Forward, Act Now: Training Young Researchers for Sustainability. Reshaping the Relationship Between PhD Student and Adviser,” in 5. International COPERNICUS Conference number 8, VAS Verlag für Akademische Schriften.
- Sánchez Reyes, L., McTavish, E., and Holder, M. (2021), “Using the Open Tree of Life for your Research, with R v0.9.1,” Open Tree of Life (online resources). AVailable at https://mctavishlab.github.io/R_OpenTree_tutorials/.
- Sandve, G. K., Nekrutenko, A., Taylor, J., and Hovig, E. (2013), “Ten Simple Rules for Reproducible Computational Research,” PloS Computational Biology, 9, e1003285.
- Sayres, M. A. W., Hauser, C., Sierk, M., Robic, S., Rosenwald, A. G., Smith, T. M., Triplett, E. W., Williams, J. J., Dinsdale, E., Morgan, W. R. et al. (2018), “Bioinformatics Core Competencies for Undergraduate Life Sciences Education,” PloS One, 13, e0196878.
- Selvaraj, A., Zhang, E., Porter, L., and Soosai Raj, A. G. (2021), “Live Coding: A Review of the Literature,” in Proceedings of the 26th ACM Conference on Innovation and Technology in Computer Science Education V. 1, pp. 164–170.
- Shannon, A., and Summet, V. (2015), “Live Coding in Introductory Computer Science Courses,” Journal of Computing Sciences in Colleges, 31, 158–164.
- Steele-Johnson, D., and Kalinoski, Z. T. (2014), “Error Framing Effects on Performance: Cognitive, Motivational, and Affective Pathways,” The Journal of Psychology, 148, 93–111.
- Sweller, J. (1988), “Cognitive Load During Problem Solving: Effects on Learning,” Cognitive Science, 12, 257–285.
- The Turing Way Community. (2021), The Turing Way: A handbook for reproducible, ethical and collaborative research (1.0.1). Zenodo. DOI: 10.5281/zenodo.6533831.
- University of Washington Libraries. (2022), “Teaching Reproducibility,” Available at https://guides.lib.uw.edu/research/reproducibility/teaching.
- Vadlamani, A., Kalicheti, R., and Chimalakonda, S. (2021), “Apiscanner-towards Automated Detection of Deprecated Apis in Python Libraries,” in 2021 IEEE/ACM 43rd International Conference on Software Engineering: Companion Proceedings (ICSE-Companion), IEEE, pp. 5–8.
- van der Meijden, A., Vredenburg, V. T., Sopory, A., Petirs, B., Tiwari, R., and Wake, D. B. (2002), “AmphibiaWeb: An Information System for Amphibian Conservation Biology. AnfibiosWeb: Un Sistema de Información Para la Biología de Conservación de Anfibios,” in Annual Meeting of the Society for Integrative and Comparative Biology, Anaheim, CA, US, January 02-06, 2002.
- Van Merriënboer, J. J., and Ayres, P. (2005), “Research on Cognitive Load Theory and its Design Implications for e-Learning,” Educational Technology Research and Development, 53, 5–13.
- Warner, J. R., Childs, J., Fletcher, C. L., Martin, N. D., and Kennedy, M. (2021), “Quantifying Disparities in Computing Education: Access, Participation, and Intersectionality,” in Proceedings of the 52nd ACM Technical Symposium on Computer Science Education, pp. 619–625.
- Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J. J., Appleton, G. I., Axton, M., Baak, A., Blomberg, N., Boiten, J.-W., da Silva Santos, L. B., Bourne, P., Bouwman, J., Brookes, A., Clark, T., Crosas, M., Dillo, I., Dumon, O., Edmunds, S., Evelo, C., Finkers, R., Gonzalez-Beltran, A., Gray, AJ., Groth, P., Goble, C., Grethe, J., Heringa, J., ’t Hoen, P.A., Hooft, R., Kuhn, T., Kok, R., Kok, J., Lusher, S., Martone, M., Mons, A., Packer, A., Persson, B., Rocca-Serra, P., Roos, M., van Schaik, R., Sansone, S-A., Schultes, E., Sengstag, T., Slater, T., Strawn, G., Swertz, M., Thompson, M., and van der Lei, J. (2016), “The FAIR Guiding Principles for Scientific Data Management and Stewardship,” Scientific Data, 3, 1–9.
- Williams, J. J., Drew, J. C., Galindo-Gonzalez, S., Robic, S., Dinsdale, E., Morgan, W. R., Triplett, E. W., Burnette III, J. M., Donovan, S. S., Fowlks, E. R., Goodman, A. L., Grandgenett, N. F., Goller, C. C., Hauser, C., Jungck, J. R., Newman, J. D., Pearson, W. R., Ryder, E. F., Sierk, M., Smith, T. M., Tosado-Acevedo, R., Tapprich, W., Tobin, T. C., Toro-Martínez, A., R. Welch, L. R., Wilson, M. A., Ebenbach, D., McWilliams, M., Rosenwald, A. G., and Pauley, M. A. (2019), “Barriers to Integration of Bioinformatics Into Undergraduate Life Sciences Education: a National Study of US Life Sciences Faculty Uncover Significant Barriers to Integrating Bioinformatics Into Undergraduate Instruction,” PloS One, 14, e0224288.
- Wilson, G. (2006), “Software Carpentry: Getting Scientists to Write Better Code by Making Them More Productive,” Computing in Science & Engineering, 8, 66–69.
- Wilson, G (2016), “Software Carpentry: Workshop Template v2016.06.” Available at https://github.com/carpentries/workshop-template.
- Wilson, G (2019), Teaching Tech Together: How to Make your Lessons Work and Build a Teaching Community Around Them, Boca Raton, FL: CRC Press.
- Wilson, G (2022), “The Carpentries,” Website. Available at http://software-carpentry.org.
- Wilson, G., Bryan, J., Cranston, K., Kitzes, J., Nederbragt, L., and Teal, T. K. (2017), “Good Enough Practices in Scientific Computing,” PloS Computational Biology, 13, e1005510.
- Wright, A. M., Schwartz, R. S., Oaks, J. R., Newman, C. E., and Flanagan, S. P. (2019), “The Why, When, and How of Computing in Biology Classrooms,” F1000Research, 8, 1854.