Abstract
T‐cell immunodeficiency is a common feature in cancer patients, which may relate to initiation and development of tumor. In expanding our previous observations in this area, we studied the repertoire of T‐cell receptor beta variable region (TRBV) and T‐cell proliferative history in CD4+ and CD8+ T cells from chronic myeloid leukemia (CML) patients. The expression and clonality analysis were performed by reverse transcription‐polymerase chain reaction (RT‐PCR) and GeneScan technique in peripheral blood mononuclear cells (PBMCs), CD4+ and CD8+ subsets of T cells. Nineteen CML cases in chronic phase were selected for this study and 17 healthy individuals served as controls. Marked restriction of TRBV repertoire was observed in both CD4+ and CD8+ T cells from CML. In most CML samples, clonally expanded T cells were identified in CD4+ and CD8+ T cells, predominantly in TRBV19 and TRBV21 (5/19) subfamilies. In conclusion, the restricted expression of TRBV subfamilies indicates the T‐cell immunodeficiency in CML patients; however, clonally expanded T cells suggest a specific immune response to leukemia associated antigens.
Introduction
Chronic myeloid leukemia (CML), with an incidence of 1·5/100 000 population, represents 15% of newly diagnosed leukemia cases in adults in China. The prognosis in CML improved markedly after introduction of ABL tyrosine kinase inhibitors (imatinib mesylate and its derivatives); however, many patients still die due to ABL mutation‐related drug resistance and blast crisis.Citation1 Therefore further studies are needed in order to better understand the disease and improve the patient outcome. T‐cell immunodeficiency may play an important role in tumor progression, facilitating the expansion of the malignant clone,Citation2,Citation3 although the interaction between the tumor and the immune system is still not completely understood.
Most circulating mature T cells use the alpha/beta heterodimeric T‐cell receptor (TCR) for specific recognition of antigenic peptides in context of major histocompatibility complex molecules. T‐cell differentiation in the thymus is characterized by a hierarchical order of rearrangement steps in the TCR genes, resulting in the joining of one of multiple variable (V), diversity (D), and joining (J) gene segments. In addition to the combinatorial joining of the V(D)J‐gene segments, the diversity of the TCR is further increased by random deletions and insertion of nucleotides between the V and J regions. This results in each differentiating T‐cell expressing unique TCR on the surface. The TCR beta locus (TRB) contains at least 64 functional V genes (T‐cell receptor beta variable region – TRBV) subdivided into 24 families. The portion of the TCR mainly responsible for the specific interaction with the antigenic peptide is the third complementarity‐determining region (CDR3) of the variable domain, including the hypervariable V(D)J junctional region. Specific recognition of an antigen results in a clonal expansion of T cells expressing unique TRBV genes with characteristic CDR3 size.Citation4 Analysis of TRBV repertoire was used to identify the distribution of TRBV subfamilies and the global profile of T‐cell subfamilies in host, while analysis of TRB CDR3 size distribution has been used to define the degree of T‐cell clonality in complex cell populations.Citation5 Oligoclonal T cells have been identified among tumor‐infiltrating T cells and peripheral blood T cells in some malignancies and in autoimmune diseases.Citation6,Citation7 Several studies on TRBV repertoire have shown that skewed expression of TRBV subfamilies is a common feature in leukemia patientsCitation8–Citation11 and clonally expanded T cells, with restricted TRBV usage, recognize tumor cells in patients with malignant disease.Citation12,Citation13
However, little data exists regarding the proliferative history of T cells in myeloid leukemia patients. Our previous study showed skewed TRBV repertoire in peripheral blood mononuclear cells (PBMCs) from 20 CML cases.Citation14 Since the high number of leukaemic cells in the blood might influence the results, in the present study, in order to more precisely characterize the immune status in CML, we analyzed the TRBV repertoire in CD4+ and CD8+ T cells from CML patients.
Materials and Methods
Samples
Nineteen newly diagnosed chronic phase CML patients, 14 males and 5 females (15–73 years old; median age: 30 years), were included in this study. BCR‐ABL fusion gene was detected in all samples by reverse transcription‐polymerase chain reaction (RT‐PCR). Seventeen healthy individuals, 6 males and 11 females (25–51 years old, median age: 28 years), served as controls. All the procedures were conducted according to the guidelines of the Medical Ethics Committees of the Health Bureau of Guangdong Province of China. The clinical data of the patients are listed in .
Table 1. Clinical data of CML patients
Mononuclear cell isolation
PBMCs were isolated from CML patients and healthy individuals by Ficoll–Hypaque gradient centrifugation.
CD3+ cell determination
CD3+ T‐cell percentage from PBMCs was determined by indirect immune fluorescent analysis. The periodate‐lysine‐paraformaldehyde‐fixed cytospin preparations were incubated with 200 μg/ml of murine anti‐CD3 monoclonal antibodies (Boster Biological Technology Ltd, Wuhan, China), washed and incubated with 1∶50 dilution of fluorescein labeled goat anti‐mouse Ig (Boster Biological Technology Ltd). The slides were counterstained with Mayer’s hematoxylin for 30 minutes. All slides were blindly evaluated using the fluorescent microscope (Nikon WFX‐II; Nikon Ltd, Tokyo, Japan); 200 cells were counted.
T‐cell sorting
The CD4+ and CD8+ T cells from PBMCs were sorted using CD4 and CD8 monoclonal antibody and MACS® magnetic cell sorting technique (Miltenyi Biotec, Bergisch Gladbach, Germany).
RNA isolation and cDNA synthesis
Total RNA was extracted from PBMCs and sorted T‐cell subsets using Trizol reagent according to the manufacturer’s recommendations (Trizol®; Invitrogen, Carlsbad, CA, USA). The quality of RNA was analyzed in 0·8% agarose gel electrophoresis and stained with ethidium bromide. The mRNA was reversely transcribed into the first single‐strand cDNA using random hexamer primers and reverse transcriptase (SuperScript® III; Invitrogen). The quality of cDNA was determined by RT‐PCR for the beta2 microglubin gene amplification.
PCR
Twenty‐four TRBV subfamily genes were amplified by RT‐PCR using 24 TRBV subfamily specific 5′ primers and a common 3′ TRBC specific primer.Citation15 PCR was performed as described by Puisieux et al.Citation7 and our previous study.Citation9 The 25 μl reaction mixture contained 1 μl cDNA, 0·5 μM sense primer and antisense primers, 0·1 mM dNTP, 1·5 mM MgCl2, ×1 PCR buffer and 1·25 U Taq polymerase (GoTaq® Flexi DNA polymerase; Promega, Madison, WI, USA). The amplification was performed in a DNA thermal cycler (Biometra, Goettingen, Germany). After 3 minutes’ denaturation at 94°C, 40 PCR cycles were performed, each cycle consisting of 94°C for 1 minute, 60°C for 1 minute, 72°C for 1 minute, and a final 7 minutes’ elongation at 72°C. Subsequently, 2 μl of the PCR products were subjected to a cycle of runoff reaction with fluorophore‐labeled TRBC‐FAM (TIB MOLBIOL GmbH, Berlin, Germany).Citation9
Genescan clonality analysis
Two microliters of FAM‐labeled runoff PCR products were heat denatured with 9·5 μl Hi‐Di formamide (ABI, Foster City, CA, USA) and 0·5 μl of size standards (GENESCAN™‐500‐LIZ™; ABI) at 94°C for 4 minutes, and loaded on the 3100 Performance Optimized Polymer‐4 gel (3100 POP‐4™; ABI). The amplicons were size separated by electrophoresis in the ABI 3100 DNA sequencer and analyzed using the GeneScan software, as described previously.Citation7,Citation9
Statistical analysis
Univariate analyses were done using the Mann–Whitney test to compare means of detectable TRBV subfamilies between PBMCs, CD4+ and CD8+ T cells groups from CML. The chi square test was used to compare the frequency of TRBV subfamily expression in CML and healthy control groups. Pearson correlation and linear regression analysis were used to estimate the correlation between age and TRBV subfamily number in PBMCs, CD4+ or CD8+ T‐cell numbers. Spearman’s correlation analysis was used to estimate the correlation between the percentage CD3+ T cells or blast+promyelocyte cells in PBMCs and the TRBV subfamily number in PBMCs, CD4+ or CD8+ T‐cell numbers, as well as the numbers of clonal‐expanded T cells in PBMCs, CD4+ and CD8+ T cells.
Results
Restricted expression pattern of 24 TRBV subfamily genes in PBMCs, and sorted CD4+ and CD8+ T cells from CML patients
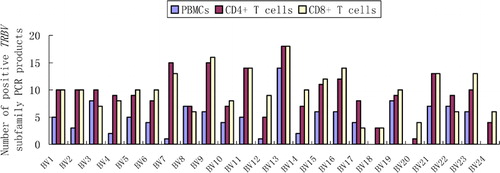
In the blood samples of healthy individuals all TRBV subfamily‐specific RT‐PCR reactions yielded positive results, whereas all 19 CML patients showed a significantly skewed blood T‐cell repertoire; 0–12 (5·63±4·11) of the 24 TRBV subfamilies were detected in PBMCs (P<0·0001). More TRBV subfamilies were detected in sorted CD4+ and CD8+ T cells (10·95±4·61 and 12·11±4·43 respectively) of CML cases, but it was again significantly lower than in corresponding T‐cell subsets from healthy individuals, which showed complete TRBV repertoire in all cases (for CD4+ T cells P<0·0001 and for CD8+ T cells P<0·0001) (–).
Figure 1. The frequency of TRBV subfamilies in PBMCs, CD4+ and CD8+ T cells from 19 CML patients. TRBV13 were used most frequently.
Clonal expansion of TRBV subfamily T cells in CD4+ and CD8+ cells from CML patients
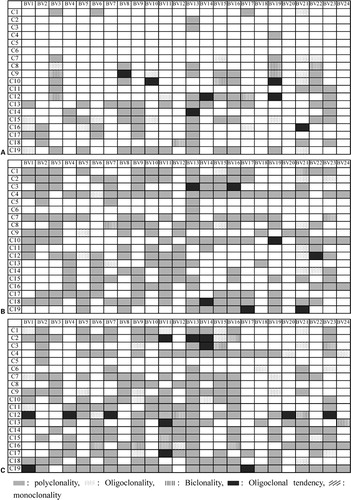
In healthy controls, CDR3 size analysis by GeneScan technique showed multipeak Gaussian distribution pattern of intensities in almost all TRBV PCR products, indicating polyclonal T‐cell repertoire. In the majority of CML patients, a part of the PCR products displayed a dominant peak corresponding to clonally expanded T cells. Clonally expanded T cells could be found in both CD4+ and CD8+ T cells in different TRBV subfamilies. Oligoclonality was most frequent in TRBV19 and TRBV21 families (5/19) (). Expansions of the TRBV genes were significantly more frequent in CML patients than in healthy controls (P = 0·015 in PBMC group, P = 0·0007 in CD4+ T‐cell group, P = 0·0001 in CD8+ T‐cell group). The clonal pattern of TRBV subfamilies in PBMCs, CD4+ and CD8+ T cells from 19 CML cases is shown in .
Correlation between the detectable TRBV numbers and the age, percentage of CD3+ T cells, blast and promyelocyte cells in PBMCs
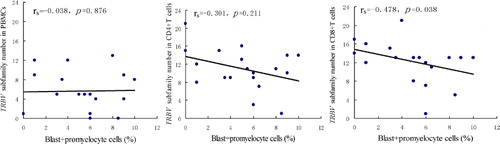
Correlation analysis was performed among the age of patients, the percentage of CD3+ cells in PBMCs from patients, the percentage of blast and promyelocyte cells in PBMCs from patients, TRBV subfamily number in PBMCs, CD4+ and CD8+ T cells. No statistically significant correlation was found between the age and the numbers of TRBV subfamilies in PBMCs, CD4+ or CD8+ T cells (R = 0·270, P = 0·264; R = 0·099, P = 0·686; and R = 0·043, P = 0·860, respectively). By Spearman’s correlation analysis, there was no significant correlation between the percentage of CD3+ T cells in PBMCs and the TRBV subfamily number in PBMCs, CD4+ or CD8+ T cells (rs = −0·029, P = 0·905; rs = 0·034, P = 0·889 and rs = 0·156, P = 0·524, respectively). The similar results were found between TRBV subfamily number in PBMCs and TRBV subfamily number in CD4+ or CD8+ T cells (rs = 0·033, P = 0·895 and rs = 0·408, P = 0·083, respectively). Significant negative correlation was observed between the percentage of blast+promyelocyte cells in PBMCs and the TRBV subfamily number in CD8+ T cells (rs = −0·478, P = 0·038) (); however, no significant correlation was found between the percentage of blast+promyelocyte cells in PBMCs and the TRBV subfamily number in PBMCs (rs = −0·038, P = 0·876) or CD4+ T cells (rs = −0·301, P = 0·211). And there was not significant correlation between the age, the percentage of CD3+ T cells, the percentage of blast+promyelocyte cells and the numbers of clonal‐expanded T cells in PBMCs, CD4+ and CD8+ T cells (data not shown).
Discussion
Understanding the mechanisms of the T‐cell immune response against tumor cells in patients with leukemia may help to develop strategies involving leukemia immunotherapy. In the first instance, an understanding of T‐cell repertoire of untreated patients is required. In healthy individuals, diverse TRBV repertoire and polyclonal CDR3 size characteristic for T cells, indicating the random rearrangement of TRBV genes in thymus.Citation8,Citation16 There have been a few attempts to characterize the TRBV usage in different types of leukemia and most have reported restricted TRBV repertoire and oligoclonal expansion of T cells.Citation8–Citation10,Citation15,Citation17–Citation21 CML is characterized by the t(9;22) translocation, in which the normal bcr and abl genes are fused. It has been demonstrated that bcr–abl fusion proteins are CML‐specific antigens, which can elicit specific anti‐CML cytotoxic T cells. Our previous study showed that the skewed repertoire of TRBV and clonal expansion of T cells are frequently detected in PBMCs from CML patients.Citation9 The restricted repertoire of T cells might be due to acquired immune deficiencies by indirect inhibition from malignant cells, leading to suppression of the proliferation of other T‐cell subfamilies; in the present study, it was showed that significant negative correlation was observed only between the percentage of blast+promyelocyte cells in PBMCs and the TRBV subfamily number in CD8+ T cells, it may explain partly the reason, or it might be due to clonal expansion in response to the leukemia associated antigens, e.g. bcr–abl fusion protein.Citation9,Citation22
Since the high number of CML cells in the blood might have influenced the results, it was thought that the restricted expression and clonal TRBV repertoire might be due to the limited number of T cells in PBMCs from patients. In the present study, in order to more precisely characterize the immune status in CML, we analyzed the TRBV repertoire in sorted T‐cell subsets, CD4+ and CD8+ T cells from 19 cases with CML. In PBMC less than 50% of TRBV subfamilies could be detected, which was similar to previously reported restricted pattern of TRBV expression.Citation23 Actually, in sorted CD4+ and CD8+ T cells more TRBV subfamilies were observed, but the restricted expression of TRBV repertoire was still a common feature in CML patients. These new data indicate that restricted TRBV repertoire is not due to high leukemic cell burden in peripheral blood but is an intrinsic feature of T cells in CML patients.
In addition to the restricted TRBV repertoire, skewed CDR3 profile was found in about half (9/19) of PBMC and in the vast majority (18/19) of sorted CD4+ or CD8+ T cells from CML patients. These observations, especially those made in sorted T cells, indicate clonal expansion of T cells, which might be due to response to leukemia‐associated antigens.Citation9 Appearance of T‐cell clones reacting specifically with the autologous leukemia cells was reported in B‐CLL.Citation11.Citation18 Patients with such clonal‐expanded T cells seemed to have better prognosis.Citation24
In conclusion, this is, to our knowledge, first characterization of the TRBV repertoire and clonality in sorted CD4+ and CD8+ T cells from CML patients. The restricted expression of TRBV subfamilies indicate the T‐cell immunodeficiency at different degree in different patients; however, the preferential usage and clonal expansion of TRBV subfamily T cells indicate that despite T‐cell immunodeficiency, CML patients have the ability for specific immune response to leukemia associated antigens.
The authors thank Dr John Yeuk‐Hon Chan for critical reading of this manuscript. This study was sponsored by grants from National ‘863’ project (No. 2006AA02Z114), National Natural Science Foundation of China (No. 39870358, 30270579) and Natural Science Foundation of Guangdong Province (No. 05103293, 9251063201000001).
References
- Jabbour E, Cortes J, Kantarjian H. Treatment selection after imatinib resistance in chronic myeloid leukemia. Target Oncol 2009;4:3–10.
- Hadden JW. Immunodeficiency and cancer: prospects for correction. Int Immunopharmacol 2003;3:1061–71.
- Costello RT, Rey J, Fauriat C, Gastaut JA, Olive D. New approaches in the immunotherapy of haematological malignancies. Eur J Haematol 2003;70:333–45.
- Pannetier C, Even J, Kourilsky P. T-cell repertoire diversity and clonal expansions in normal and clinical samples. Immunol Today 1995;16:176–81.
- Sensi M, Parmiani G. Analysis of TCR usage in human tumors: a new tool for assessing tumor-specific immune response. Immunol Today 1995;16:588–95.
- Lozeron P, Chabas D, Duprey B, Lyon-Caen O, Liblau R. T cell receptor V beta 5 and V beta 17 clonal diversity in cerebrospinal fluid and peripheral blood lymphocytes of multiple sclerosis patients. Mult Scler 1998;4:154–61.
- Puisieux I, Even J, Pannetier C, Jotereau F, Favrot M, Kourilsky P. Oligoclonality of tumor-infiltrating lymphocytes from human melanomas. J Immunol 1994;153:2807–18.
- Li Y. Leukemia associated clonal expansion TCR Vβ subfamily T cells. Hematology 2003;8:375–84.
- Li Y, Yang L, Chen S, Zhang Y, Wu X. The TCR Vbeta repertoire usage of T-cells from cord blood induced by chronic myelogenous leukemia associated antigen. Hematology 2005;10:387–92.
- Rezvany M, Jeddi-Tehrani M, Osterborg A, Kimby E, Wigzell H, Mellstedt H. Oligoclonal TCR BV gene usage in B-cell chronic lymphocytic leukemia: major perturbations are preferentially seen within the CD4 T-cell subset. Blood 1999;94:1063–9.
- Rezvany MR, Jeddi-Tehrani M, Wigzell H, Osterborg A, Mellstedt H. Leukemia-associated monoclonal and oligoclonal TCR-BV use in patients with B-cell chronic lymphocytic leukemia. Blood 2003;101:1063–70.
- thor Straten P, Guldberg P, Gronbeak K, Hansen MR, Kirkin AF, Seremet T, et al.. In situ T cell responses against melanoma comprise high numbers of locally expanded T cell clonatypes. J Immunol 1999;163:443–7.
- Mami-Chouaib F, Echchakir H, Dorothée G, Vergnon I, Chouaib S. Antitumor cytotoxic T-lymphocyte response in human lung carcinoma: identification of a tumor-associated antigen. Immunol Rev 2002;188:114–21.
- Geng SX, Li YQ, Chen SH, Yang LJ, Yin QS, Wu XL, et al.. Peripheral blood naive T cell level and its T cell receptor Vbeta repertoire usage profile in patients with chronic myelogenous leukemia.Zhonghua Xue Ye Xue Za Zhi 2005;26:413–6(Chinese).
- Li Y, Chen S, Yang L, Yin Q, Geng S, Wu X, et al.. TRAV and TRBV repertoire, clonality and the proliferative history of umbilical cord blood T cells. Transplant Immunol 2007;18:151–8.
- Gorski J, Yassai M, Zhu X, Kissela B, Kissella B. Circulating T cell repertoire complexity in normal individuals and bone marrow recipients analyzed by CDR3 size spectratyping. J Immunol 1994;152:5109–19.
- Assaf C, Hummel M, Dippel E, Goerdt S, Muller HH, Anagnostopoulos I, et al.. High detection rate of T-cell receptor beta chain rearrangements in T-cell lymphoproliferations by family specific polymerase chain reaction in combination with the GeneScan technique and DNA sequencing. Blood 2000;96:640–6.
- Farace F, Orlanducci F, Dietrich PY, Gaudin C, Angevin E, Courtier MH, et al.. T cell repertoire in patients with B chronic lymphocytic leukemia. Evidence for multiple in vivo T cell clonal expansions. J Immunol 1994;153:4281–90.
- Coleman S, Perera P, Fisher J, Hoy T, Burnett AK, Lim SH. Abnormal TCR V beta repertoire in patients with chronic myeloid leukaemia. Br J Haematol 1995;90:358–63.
- Serrano D, Monteiro J, Allen SL, Kolitz J, Schulman P, Lichtman SM, et al.. Clonal expansion within the CD4+CD57+ and CD8+CD57+ T cell subsets in chronic lymphocytic leukemia. J Immunol 1997;158:1482–9.
- Kluin-Nelemans JC, Kester MG, Melenhorst JJ, Landegent JE, van de Corput L, Willemze R, et al.. Persistent clonal excess and skewed T-cell repertoire in T cells from patients with hairy cell leukemia. Blood 1996;87:3795–802.
- Alatrakchi N, Farace F, Frau E, Carde P, Munck JN, Triebel F. T-cell clonal expansion in patients with B-cell lymphoproliferative disorders. J Immunother 1998;21:363–70.
- Li YQ, Yang LJ, Chen SH, Zhang YP, Zhang XL, Luo GX. TCR Vβ repertoire usage and clonal expansion of T cells in chronic myelogenous leukemia. Chin Med J 2004;117:840–3.
- Brown RD, Yuen E, Nelson M, Gibson J, Joshua D. The prognostic significance of T cell receptor beta gene rearrangements and idotype-reactive T cells in multiple myeloma. Leukemia 1997;11:1312–7.