Abstract
Micro RNAs are a class of small non-coding RNAs which has been recently shown to play a crucial role in major cellular processes such as development and differentiation through post-transcriptional regulation. The role of these epigenetic elements has also been demonstrated in hematopoietic lineage differentiation and there is a large body of evidence that miR-424 is responsible for monocyte differentiation. Our goal was to examine the effect of miR-424 over-expression on defeating the maturation blockage in monoblastic cell line U937. The permanent over-expression of miR-424 was established using a retroviral vector construct containing the precursor of miR-424 sequence. Induction of differentiation process was monitored by assaying changes in cell morphology, and expression of cell surface markers using light microscopy, quantitative RT-PCR, and flow cytometry for monocyte markers such as CD11b and CD14. The cells showed monocytic characteristics 14 days after transduction, and CD11b and CD14 expression were significantly increased, confirmed by flow cytometry QRT-PCR and RT-PCR results. In conclusion, miR-424 over-expression is an effective factor in maturation of the monoblastic U937 cells and it has the ability of directing them into cells, expressing monocyte/macrophage characteristics.
Introduction
According to the World Health Organization classification of myeloid neoplasm and acute leukemia, acute myelomonocytic leukemia, acute monoblastic/monocytic leukemia, and AML with t(9;11) (p22;q23) MLLT3-MLL are considered as malignancies associated with a failure, most dominantly, in maturation and, therefore, apoptosis of progenitors in monocyte lineage.Citation1 Hence, overcoming this differentiation arrest and inducing maturation and apoptosis could result in having cells with partial normal activity and hopefully a remission.Citation2
The U937 cell line, which is originally established from a generalized histiocytic lymphoma, is a hematopoietic cell line displaying a number of features similar to monocyte progenitor cells. For this reason, the U937 cell line has been utilized as an in vitro model for the study of differentiation in hematopoietic disorders, and such malignancies mentioned above, because of its monoblastic properties.Citation3–Citation5 The U937 cell line differentiation along the monocyte/macrophage pathway has been demonstrated before.Citation6 Induction of differentiation in U937 cell line into mature and non-proliferative monocytes is feasible using several factors such as: vitamin D3, all-trans-retinoic acid, and 12-O-tetradecanoylphprbol-13-acetate.Citation5,Citation7
Recently, a class of small non-coding RNAs, known as microRNAs, has been found to play a critical role in major cellular processes such as development and differentiation through post-transcriptional regulation.Citation8,Citation9 The role of these epigenetic elements has been also demonstrated in hematopoietic lineage differentiation. Ectopic over-expression of miR-181 in mouse hematopoietic progenitors led to increased production of B-cells,Citation10 and it has also been shown that miR-223 affects granulocytic differentiation.Citation11 Similarly, there is a large body of evidence that increase in the miR-424 during monocyte/macrophage differentiation has an effect to some extent in a similar way to miR-233 in the granulocytic differentiation pathway.Citation12
In this study, we established ectopically over-expression of miR-424 in the U937 cell line, using a retroviral expression system to investigate the capability of miR-424 in overcoming the blockage of differentiation, existing in the mentioned cell line.
Materials and Methods
Plasmids and vector construction
The pRETRO-424 plasmid was generated by cloning a fragment containing the pri-miR-424 (from −120 to +160 bp relative to the 5′-end of pre-miR-424) into the pRETRO SUPER plasmid which was a gift from Dr Ghazizadeh S. (Department of Oral Biology and Pathology, State University of New York at Stony Brook, New York, USA) The pri-miR-424 was obtained by a polymerase chain reaction on human genomic DNA extracted from white blood cells using blood & cell culture DNA mini kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. The primers had an overhang tail, containing cutting sites for specific restriction enzymes correlated with the multiple cloning site of the vector (forward primer: 5′-GGAAGATCTGAAGTGGCCTAGTCATxAAG-3′ and reverse primer: 5′-CCCAAGCTTCTCACTGCAGAACTGTTC-3′). The ligation of insert oligonucleotide and the vector were completed using T4 DNA ligase enzyme (Fermentas, Burlington, VT, USA) based on manufacturer’s instructions.
Virus packaging and concentration
For virus packaging, we used GP293T cell line. The mentioned cell line expresses the gag/pol packaging genes. pMDG plasmid containing vsv-g were co transfected with pRETRO-424 (as well as for pRETRO backbone) into GP293T cell line using Express-In transfection reagent according to manufacturer’s instructions (Open Biosystems, Huntsville, AL, USA). The supernatant containing the produced viruses (RV-424) were collected after 24, 48, and 72 hours after transfection (the early collected supernatant were kept at 4°C during the next 48 hours) and went through the concentration process which included centrifuging in 21 000 rev/min or 40 000g at 4°C for 2 and a half hours. The concentrated supernatant was aliquoted and stored at −70°C. For titration of functional virus particles 293T cells were used as a host for packaged viruses. In order to determine the virus copy number, 48 hours after transduction of 293T cells with RV-424, genomic DNA of those cells was extracted and purified using blood and cell culture DNA mini kit (Qiagen). The primers used were specific for the puromycin resistance gene existing on the vector (forward primer: 5′-GCAGCAACAGATGGAAGG-3′ and reverse primer: 5′-GAGGTCTCCAGGAAGGC-3′). The final titration based on a standard curve and real-time PCR result using specific primers for the RV-424 construct was 10Citation9 viral particles/ml.
Cell cultures and transduction
U937 cell line was purchased from Pasteur Institute of Iran and cultured in DMEM medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Gibco) for expansion. The same medium was used for GP293T packaging cell line, during the viral packaging. For over-expression study, the U937 cells were transferred into a 12-well plate containing IMDM medium (Gibco) supplemented with 10% fetal bovine serum for 2 weeks. All the media were contained penicillin, streptomycin (Gibco) and L-glutamine (Gibco) and the incubation conditions were 37°C and 5% CO2. Cells were seeded at density of 1×105 cells/well (untreated control) and 2×105 cells/well (RV-424- and empty vector-treated). Several concentration and administration protocols were tested (data not shown), but the following conditions were chosen. The U937 cells were transduced with a multiplicity of infection (MOI) of 10. The virus was added to U937 cells and gently mixed on a bio shaker 3D for 6 hours at 37°C and 5% CO2. Twenty-four hours after transduction, the media were refreshed. The cells were collected, and analyzed for expression of cell surface markers at days 7 and 14.
Differentiation analysis
The U937 cells were collected at days 7 and 14 after transduction. Expressions of surface monocytic markers were analyzed using flow cytometry and quantitative RT-PCR. Expression of CD11b, CD14 and CD64 were analyzed in flow cytometric assays and mRNA expression of CD11b and CD14 detected by quantitative RT-PCR.
Flow cytometry
For surface markers detection, purified PE-conjugated antibody against CD11b (DAKO Denmark, Glostrup, Denmark) and FITC-conjugated antibody against CD14 (DAKO Denmark) were used according to the manufacturer’s instructions. FITC-conjugated and PE-conjugated mouse IgG1 antibody (DAKO Denmark) was used as an isotype-matched control. After labeling, the cells were washed and suspended in 1% (w/v) paraformaldehyde. The cells were processed in a FACScan flow cytometer Partec PAS III (Partec, Munster, Germany) equipped with an argon and red diode laser. FloMax 2·4 software (Partec FloMax operating and analyzing software; Partec) was used to create the histograms. Cells were gated out with a forward versus side scatter window.
RNA extraction and cDNA synthesis
Total RNA, extracted using Qiazol reagent (Qiagen) according to manufacturer’s instructions, cDNA synthesis were carried out using cDNA synthesis kit (Fermentas) based on manufacturer’s instruction. mRNA quantification was performed by real-time PCR with Fermentas SYBR green mastermix (Fermentas), according to the manufacturer’s instructions in a Rotor-Gene 6000 system (Corbett, Concorde, NSW, Australia). The following oligonucleotides were used as detection primers: CD11b Fwd, 5′-TCCAAAACACGGGGACCTATC-3′ and Rev, 5′-TCCTCGAACACGACCACCT-3′, and CD14 Fwd, 5′-AGACTTATCGACCATGGAGC-3′ and Rev, 5′-TCTACTGCAGACACACACTG-3′. Delta-delta Ct values were normalized with those obtained from the amplification of the endogenous GAPDH mRNA in the same manner as others.
Results
Cell morphology
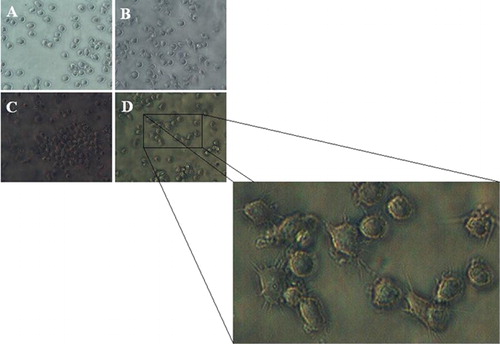
shows four representative micrographs of U937 cells at day 14 after transduction. The images are respectively grown in virus free condition (), transduced with backbone of the vector (), and transduced with RV-424 (). The culture illustrated in shows roundish and floating cells that still proliferate. Cells of the culture shown in have the same morphological features as the control group A which is a evidence that backbone of the vector, by itself, does not stimulate the monocytic differentiation. The cells in cultures display the differentiated cells with pseudo-pods and flatten macrophage-like morphology, adherent to the substrate.
Transcription of monocyte/macrophage markers
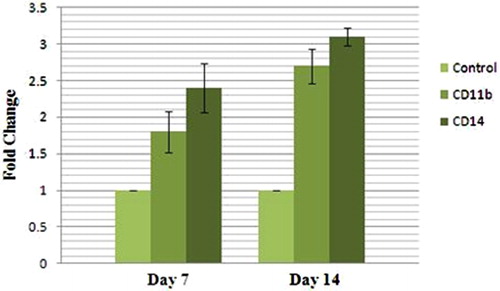
In order to assure that differentiation has taken place, the mRNA transcript of two monocytic/macrophage markers were measured by quantitative RT-PCR. CD11b is a surface marker of granulocytic and monocytic cell differentiation, whereas CD14 specifically recognizes cells of the monocytic lineage. shows that CD11b-positive cells almost doubled during the 14 days (1·8±0·28-fold at day 7, and 2·7±0·24-fold at day 14), whereas those positive for CD14 reached an about three-fold increase at day 14 after transduction (2·4±0·34-fold at day 7, and 3·1±0·12-fold at day 14). These data indicate that ectopic over-expression of miR-424 has an effect on monocyte/macrophage differentiation and miR-424 is capable of overcoming the differentiation arrest existing on the U937 cells.
Surface marker expression
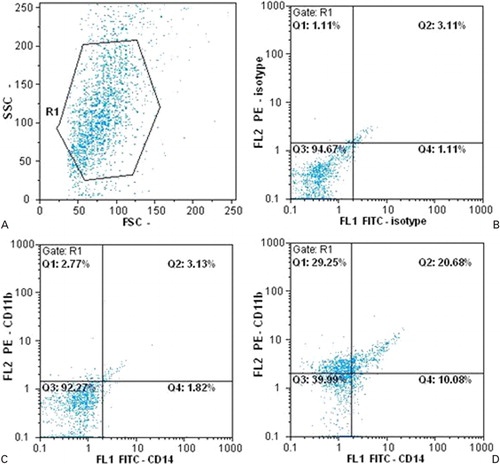
The expressions of CD11b and CD14 on cell surface were detected by flow cytometric assay at day 14 after transduction. The results show that cells expressing the CD11b at day 14 are increased in comparison with the group of cells that only transduced with the backbone of the vector (2·77% in the control group and 29·25% in the treated group).
In the same way but with different values, there is an increase in monocyte-specific marker CD14 in the group of cells transduced with the RV-424, compared with the control culture (1·82% in the control group and 10·08% in the RV-424 treated group). Therefore, CD11b-expressing cells had an about 10-fold increase, whereas CD14-positive cells show a five-fold increase compared with the control group. However, double positive cells (cells that express CD11b and CD14 simultaneously) have increased from 3·13% in group of cells transduced with empty vector to 20·68% in the RV-424 group 14 days after transduction, which means that there was almost seven-fold increase in double positive cells in the group that was transduced with RV-424 ().
Discussion
As expected, over-expression of miR-424 in the U937 cell line leads to induction of differentiation into monocyte/macrophage-like cells with regard to the following criteria: morphological characteristics, cell growth inhibition, and expression of cell surface markers.
As mentioned before, microRNAs are small, non-coding, and extremely conserved RNA molecules which regulate expression of genes at post-transcriptional stage by binding to the 3′-UTR of the target mRNAs.Citation13–Citation15 Many studies have demonstrated the importance of individual microRNAs to affect physiological processes, such as hematopoietic cell development and differentiation.Citation15–Citation18 Additionally, it has been shown that several microRNAs are broadly expressed in hematopoietic cells, and the altered expression of them (e.g. by chromosomal translocations) is associated with leukemia.Citation15,Citation19,Citation20 Besides that, there are several reports about an individual micro RNA (miR-424) which promote monocytic differentiation, and it seems that miR-424 is capable of overcoming the uncontrolled proliferation of progenitors, in some monocytic malignancies. The effect of over-expression of mentioned micro-RNA has been studied on a number of cell lines such as THP-1 cells (an M5-AML containing the MLL-MLLT3 fusion),Citation21 and NB4 cells (a genuine human promyelocytic cell line derived from the bone marrow of an APL patient).Citation12 We show that miR-424 can also induce differentiation of the U937 cells into an adherent and aggregated monocyte-like phenotype, comparable to the U937 cells exposed to 12-O-tetradecanoylphprbol-13-acetate.Citation5 The induced differentiated cells also showed several changes on mRNA level of monocyte/macrophage associated genes, CD11b and CD14 ().
We also show that CD11b and specially CD14 proteins expression on cell surface, increased after monocyte differentiation induction using the miR-424. Our data about expression of CD14 are similar to the result from experiments on CD34+ cellsCitation12 (peak of expression after almost 2 weeks), but not exactly the same response as THP-1, considering that miR-424 induces monocyte/macrophage differentiation by targeting NFI-A in CD34+ stem cells, and the fact that NFI-A does not expressed in THP-1 cells, assuming that in THP-1 cells, other targets and regulators are involved.Citation12,Citation21 Taking it all together, we suggest that the U937 cells are probably a better model for studying monocyte/macrophage differentiation in this matter.
In conclusion, we demonstrated that differentiation block on U937 cells could be bypassed using miR-424 differentiation induction. Our future prospect is to investigate the role of this individual micro-RNA during monocyte/macrophage differentiation in a more primary hematopoietic stem cell along with several other miRNAs which seem to have effect on above cell development.
This work was supported by the Iranian Council of Stem Cell Technology, Faculty of Medical Sciences of Tarbiat Modares University, and the Stem Cell Technology Research Center, Tehran, Iran.
References
- Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al.. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 2009;114:937–51.
- Ferrara FF, Fazi F, Bianchini A, Padula F, Gelmetti V, Minucci S, et al.. Histone deacetylase-targeted treatment restores retinoic acid signaling and differentiation in acute myeloid leukemia. Cancer Res 2001;61:2–7.
- Harris P, Ralph P. Human leukemic models of myelomonocytic development: a review of the HL-60 and U937 cell lines. J Leukoc Biol 1985;37:407–22.
- Sundstrom C, Nilsson K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int J Cancer 1976;17:565–77.
- Minafra L, Di Cara G, Albanese NN, Cancemi P. Proteomic differentiation pattern in the U937 cell line. Leuk Res 2010;35:226–36.
- Caligo MA, Cipollini G, Petrini M, Valentini P, Bevilacqua G. Down regulation of NM23.H1, NM23.H2 and c-myc genes during differentiation induced by 1,25 dihydroxyvitamin D3. Leuk Res 1996;20:161–7.
- Stockbauer P, Malaskova V, Soucek J, Chudomel V. Differentiation of human myeloid leukemia cell lines induced by tumor-promoting phorbol ester (TPA). I. Changes of the morphology, cytochemistry and the surface differentiation antigens analyzed with monoclonal antibodies. Neoplasma 1983;30:257–72.
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al.. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 2005;102:13944–9.
- Kluiver J, Kroesen BJ, Poppema S, van den Berg A. The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia 2006;20:1931–6.
- Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005;11:241–7.
- Fazi F, Rosa A, Fatica A, Gelmetti V, de Marchis ML, Nervi C, et al.. A minicircuitry comprised of MicroRNA-223 and transcription factors NFI-A and C/EBPα regulates human granulopoiesis. Cell 2005;123:819–31.
- Rosa A, Ballarino M, Sorrentino A, Sthandier O, de Angelis FG, Marchioni M, et al.. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci USA 2007;104:19849–54.
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science 2001;294:853–8.
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001;294:862–4.
- Hughes PJ, Marcinkowska E, Gocek E, Studzinski GP, Brown G. Vitamin D3-driven signals for myeloid cell differentiation — implications for differentiation therapy. Leuk Res 2009;34:553–65.
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004;303:83–6.
- Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood 2006;108:3646–53.
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, et al.. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008;451:1125–9.
- Fabbri M, Garzon R, Andreeff M, Kantarjian HM, Garcia-Manero G, Calin GA. MicroRNAs and noncoding RNAs in hematological malignancies: molecular, clinical and therapeutic implications. Leukemia 2008;22:1095–105.
- Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell 2008;133:217–22.
- Forrest AR, Kanamori-Katayama M, Tomaru Y, Lassmann T, Ninomiya N, Takahashi Y, et al.. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia 2009;24:460–6.