Abstract
Objective: This study aims to investigate the association between the polymorphisms in DNA repair genes (XPD, XRCC1, and XRCC4) and clinical parameters in patients with multiple myeloma (MM), their effects on prognosis and their roles in susceptibility to MM.
Patients and methods: Sixty patients, diagnosed with MM and 70 individuals as the healthy control group were included in the study. Gene polymorphisms were detected with the polymerase chain reaction and/or polymerase chain reaction–restriction fragment length polymorphism method. When the genotype frequencies of XPD (Llys751Gln) and XRCC1 (Arg399Gln) genes were examined in the patient and control groups, no significant difference was detected, while a significant association was found in XRCC4 (VNTR in intron 3 and G-1394T) polymorphisms. A significant association was found in the MM patients group for AA genotype and event-free survival (EFS) in terms of XPD (751) gene polymorphism (P = 0·047). When VNTR intron 3 polymorphism was compared for genotype frequency, DD genotype was found to be significantly low (P = 0·012) in the patient group, whereas GG and TT genotypes were found to be significantly lower in the patient group for the genotype frequency XRCC4 (G-1394T) polymorphism when compared to the control group (P = 0·015, P = 0·010, respectively).
Results: These data provide support for the hypothesis that a common variation in the genes encoding XRCC4 DNA repair proteins may contribute to susceptibility to myeloma. These findings require further validation in independent populations.
Introduction
Multiple myeloma (MM) is a B-cell lymphoproliferative disease in which, in general, progessive immunoglobulin generation with increased malignant plasma cells in the bone marrow is seen, and is generally characterized with osteolytic lesions, hypercalcemia, anemia, monoclonal protein increase, and renal failure.Citation1Citation1,2 It has been suggested that radiation (short-term high dose or long-term low dose), viral agents, chronic antigenic stimulus, and environmental factors have a role in MM etiology.Citation3
Two oncogenic pathways are believed to play central role in myeloma pathogenesis. First, multiple trisomies of chromosomes; 3, 5, 7, 9, 11, 15, 19 and 21 are related to disease pathogenesis. Second, primary translocation involving heavy chain locus at 14q32 and a limited number of recurrent locus are present.Citation4 These translocations represent abnormal class switch recombination which normally functions to alter immunglobuline isotype with the maturation of the immune response. Disregulated class switch recombination results in the placing of a number of proto-oncogenes and is believed to play a role in myeloma pathogenesis.Citation5
Minor damages in the DNA molecule are generally cleared by DNA repair genes. Under normal circumstances, high level DNA damages leads to cell death by stimulating apoptosis.Citation6 Xeroderma pigmentosum group D (XDP) gene is a DNA repair gene and is found in the nucleotide excision repair (NER) group. During NER, some chemically altered, mismatched or unsuitable bases are excised from the genome, and bases in correct order are replaced. Transformation of lysine in 751th codon of XDP gene to glutamine alters the response to treatment. NER mechanism mostly participates in DNA damage due to ultraviolet light and polycyclic hydrocarbonates.Citation7 X-ray repair cross-complementing group 1 and 4 (XRCC1 and XRCC4) are DNA repair genes in subgroup of the base excision repair (BER) mechanism. BER mechanism participates in DNA damage due to X rays, oxygen radicals and alkylating agents, while mismatch repair is assigned in reparation of replication failures.Citation8 In XRCC1 base damages and one-stranded DNA breaks, DNA functions by binding to damaged DNA part of poly(ADP-ribose) polymerase and DNA polymerase b, and to carboxyl of Ligase III.Citation9
Polymorphisms causing changes in DNA repair capacity have been known to hold an important place in MM etiology and the response to the treatment. Both karyotypic and evidence of DNA repair abnormalities take place in myeloma pathogenesis, and we selected three genes which are involved in DNA repair as the basis for this study. XRCC4 has also been implicated in myeloma pathogenesis.Citation10 Therefore, we investigated XPD (Lys751Gln), XRCC1 (Arg399Gln) polymorphisms and also XRCC4 (VNTR in intron 3 and G-1394T) for assessment of myeloma pathogenesis and relation to clinical parameters.
Patients and Methods
Sixty patients, consecutively diagnosed with MM between 2004 and 2008 in the Section of Hematology at the Department of Internal Medicine in the Faculty of Medicine at Gaziantep University, and 70 individuals as the healthy control group were included in the study. The diagnosis was based on clinical criteria, serum protein electrophoresis, immune electrophoresis and immune fixation of serum and urine, and bone marrow examination. The study was approved by the regional ethics committee, and informed consent was obtained from each participant before interview and blood sampling. DNA isolation was performed using 2 cc of blood sample in tubes with EDTA for the control and from bone marrow smear preparations for the patient group at diagnosis. The control group was composed of individuals with no previous a disease history and with completely normal clinical and laboratory findings. DNA isolation was performed using Invitrogen Pure Link (Catalogue Nr: K1820-01) kits in patient group, and control group. The DNA samples were stored at 20°C. Gene polymorphisms were detected with the polymerase chain reaction and/or polymerase chain reaction–restriction fragment length polymorphism methodCitation11Citation11,12 ().
Table 1. XRCC1 (A399G), XPD (−751), XRCC4 (VNTR in intron 3 and G-1394T) primer sequences, restriction enzymes and amplification conditions (on over night incubation)
The analyses of data were performed using the software SPSS for Windows (version 13·0; SPSS, Chicago, IL, USA). The statistical significance of the differences between the patient and the control groups was estimated by logistic regression analysis. Adjusted odds ratios (ORs) were calculated with a logistic regression model that controlled for gender and age and were reported at 95% confidence intervals. Differences in DNA Repair Gene XPD, XRCC1, XRCC4 (VNTR in intron 3 and 1394) allele genotype frequency between the control group and the patient groups were compared using the χ2test and, when needed, Fisher exact test was used. Hardy–Weinberg equation was used to calculate estimated genotype frequency and the experienced genotype frequency.Citation13 Survival probabilities were estimated by the Kaplan–Meier method and differences were compared using the log rank test.Citation14 The Cox stepwise regression analysis was employed to confirm significance of risk factors.Citation15 In multivariate analysis, we used eliminated variables stepwise (backward) by a significance of less than 10%. P values <0·05 were considered to indicate statistical significance.
Results
In the study, 34 (57%) of the cases were men, and 26 (43%) of them were women. The average age was 59 years (36–80 years). In the control group there were 37 (53%) men, and 33 (47%) women out of 70 healthy volunteers. The average age of the control group was 41 years (20–66 years). Results relating to XPD, XRCC1, XRCC4 gene polymorphisms of the 60 patients diagnosed with MM are shown in . Paraprotein types detected in MM patient group, stage (according to Durie–Salmon classification), Eastern Cooperative Oncology Group (ECOG) performance scale, demographic and other clinical characteristics of the patients are seen in . Eight of the patients who were assessed died (infection, heart failure, cerebrovascular accident, gastrointestinal and central nervous system bleeding) shortly after their hospitalization, and were not included in the survival assessment.
Table 2. Clinical characteristics and treatment regimens of multiple myeloma patients
When XPD (Lys751Gln) gene polymorphism results were compared in both control (P = 0·124) and patient group (P = 0·233) a deviation from Hardy Weinberg Equilibrium (HWE) was not detected. When XRCC1 (Arg399Gln) gene polymorphism results were compared in both patient (P = 0·0011) and control group (P = 0·009) a deviation from HWE was detected. When XRCC4 (VNTR in intron 3) gene polymorphism results were compared a deviation from HWE in control group (P = 0·002) was not detected while in patient group (P = 0·002) a deviation from HWE was detected. When XRCC4 (G-1394T) gene polymorphism results were compared in control group (P = 0·043) there was a deviation from HWE, but a deviation was not detected in patient group (P = 0·497). When genotype frequencies obtained from DNA repair gene polymorphisms in MM patient and control group were compared, a significant correlation in XDP (Lys751Gln) and XRCC1 (Arg399Gln) polymorphism results was not found while such a relationship was detected in XRCC4 (VNTR intron 3 and G-1394T) polymorphisms ().
Table 3. Comparison of genotype of XPD, XRCC1, XRCC4 gene polymorphisms between patients with multiple myeloma and healthy controls
Using International Myeloma Working Group Uniform Response Criteria,Citation16 initial treatment and treatment results of patients whose response evaluations are complete are seen in . In univariate analyses of MM patients, for male gender 6-years median EFS was found significant, while in patients whose IPI score was I/III, ECOG performance scale was <1/>1 and platelet number was <150×109/l/⩾150×109/l, 6-years OS was found significant (respectively P = 0·044, P = 0·037, P = 0·033) (). While in XPD (Lys751Gln) gene group AA genotype was found insignificant (P = 0·061) in comparison to AC and CC genotypes in terms of 6-years median OS, it was found significant (P = 0·047) () () in terms of EFS. However, in multivariate analyses, a significant correlation was not found in terms of male gender, IPI score, ECOG performance scale, platelet number, XDP (751) AA genotype frequency, OS and EFS. In MM patients, a significant correlation was not detected between XDP, XRCC1, XRCC4 gene polymorphisms and beta2 microglobulin, disease stage, paraprotein type, gender, lactic dehydrogenase, plasma cell ratio, seasonal relation of the disease and leukocyte number. Also there was no association of age with XDP, XRCC1, XRCC4 gene polymorphisms.
Figure 1. Kaplan–Meier plots on overall survival (OS) time according to the type of IPI risk score at diagnosis.
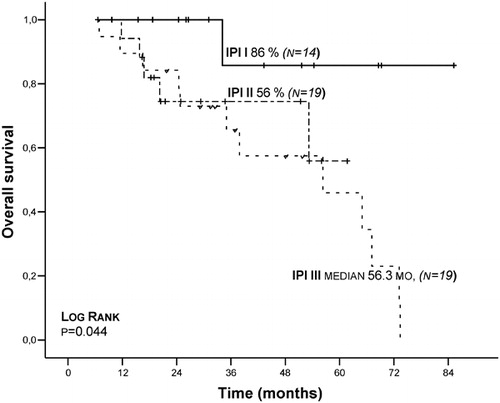
Figure 2. Kaplan–Meier plots on event-free survival (EFS) time according to the type of XPD (Lys751Gln) genotypes.
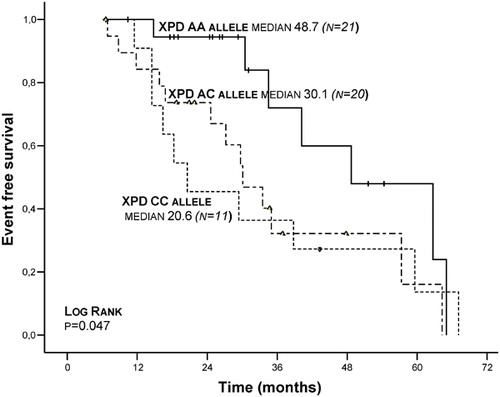
Table 4. Initial treatment modalities and treatment response of myeloma patients
Table 5. Univariate analysis (Log rank test) of XPD, XRCC1, XRCC4 − intron 3 and XRCC4 – 1394 Gene Polymorphisms in the patients with multiple myeloma
Discussion
Correlations between polymorphisms in DNA repair genes and the risk of developing various cancers using meta-analyses has been shown. Defective DNA repair has also been implicated directly as an etiological factor in hematological malignancies.Citation17,Citation18 Our study shows that in MM patients, XDP (Lys751Gln), XRCC1 (Arg399Gln), XRCC4 (VNTRintron 3 and G-1394T) gene polymorphisms functioning in DNA repair systems have no effect on treatment protocol, prognosis, and survival; however, XDP (Lys751Gln) AA genotype is significant in terms of 6-years EFS (, and ). It was found that DD genotype in XRCC4 (VNTR intron 3) and GG and TT genotype in XRCC4 (G-1394T) are significantly low in the patient group ().
In some of the studies on XPD (Lys751Gln) polymorphism, it was noted that the XPD Lys allele is associated with an increased risk for lung cancer, melanoma and basal cell carcinoma.Citation19 A correlation between XPD Gln allele and acute myeloblastic leukemia developing following chemotherapy was found, while a correlation between other prognostic factors (age, gender, WBC number, FAB class, response to treatment) and XPD (Lys751Gln) polymorphism was not detected.Citation20Citation20,21 In follicular lymphoma patients, ERCC2 and ERCC1 gene polymorphisms was not associated with the disease; however, it was stated that follicular lymphoma patients who are smoking and XRCC3 gene variations can be related to the disease.Citation22In our study, in 60 MM patients XDP (Lys751Gln) gene distribution was as following: AA: 25 (41·7%), AC: 24 (40%), CC: 11 (18·3%); a deviation from HWE was not detected in control groups. A significant disparity between MM patient group (P = 0·12) and control group (P = 0·22) was not detected in terms of genotype frequency. In our study, in XPD (Lys751Gln) gene group, AA genotype was not found significant (P = 0·061) in comparison with AC and CC genotype in terms of 6-years median OS; however, it was found significant (P = 0·047) in terms of EFS (, ). In this condition, in MM patients, XPD (Lys751Gln) AA genotype can be said to play a protective role. However, in multivariate analyses, a significant correlation was not detected in terms of XPD (751) AA genotype frequency OS and EFS.
In a study in which the relationship between XRCC1 polymorphisms (codon 194, 280, and 399) and acute lymphoblastic leukemia (ALL) risk was investigated, it was detected that only XRCC1 Gln variant allele constitutes a risk for acute lymphoblastic leukemia.Citation23 XRCC1 (A194T allele) polymorphism is associated with an insignificant increase in NHL risk, while this allele was noted to be associated with a decrease in diffuse large B-cell lymphoma. In another study, it was reported that a correlation between XRCC1 (Arg339 Gln) polymorphism and NHL subtypes did not exist.Citation24Citation24,25In our study, in 60 patients diagnosed with MM, XRCC1 (Arg399Gln) gene polymorphism distribution was as following: AA: 23 (38·3%), AG: 20 (33·3%), GG: 17 (28·4%); and in both control (P = 0·009) and patient group (P = 0·0011) a deviation from HWE was detected. As a result of the comparison of MM patient group to control group, a significant disparity between XRCC1 codon (Arg399Gln) gene polymorphisms and patient group was not found in terms of genotype frequency and clinical parameters ().
In XRCC4 G-1394T gene polymorphism, an increased correlation between T/T genotype and gastric cancer was detected, although it was explained that XRCC4 (247, intron 3 and intron 7) gene polymorphism was not associated with gastric cancer.Citation26Citation26,27In a study by Hayden et al. in which XRCC3, XRCC4, and XRCC5 polymorphisms were investigated and in which 307 MM and 263 control group patients were participated, single nucleotide polymorphisms were studied, and it was noted that A allele (P = 0·0133) in rs963248 numbered single nucleotide polymorphisms in XRCC4 gene, and AA genotype (P = 0·026) increased significantly in patient group in comparison with control group. In this case, they also reported that it increased susceptibility to MM.Citation10 In our study, out of 60 cases, XRCC4 (VNTR intron 3) polymorphism distribution was detected as: DD: 1 (1·7%), DI: 36 (60%), II: 23 (38·3%). While a deviation from HWE in control group and in the MM patient group was not detected. When patient and control group were compared in terms of DD, DI, and II genotypes, it was determined that DD genotype was found to be significantly low (P = 0·012) in patient group in comparison with healthy group. In this case, it can be implied that as DD genotype decreases, susceptibility to MM increases, and protectiveness from MM disappears as DI and II increases.
In our study, in patient group XRRC4 (G-1394T) gene polymorphism distribution was detected as: GG, 20 (33%); GT, 27 (45%); TT, 13 (21·7%). When a healthy control group and MM patient group were compared, in terms of genotype frequency GG and TT genotype were found to be significantly low in patient group in comparison with healthy control group (). We believe that this decrease in GG and TT genotype may increase the susceptibility to MM, and its high incidence in control group may be shown as a protection-creating situation against tumor.
Conclusion
In this study, XPD (Lys751Gln) and XRCC1 (Arg399Gln) gene polymorphisms functioning in BER and NER DNA repair system in MM patients were investigated. The polymorphisms of these areas were found to be significant, and we believe the genes’ different loci may be related to MM pathogenesis and these different loci should be further investigated. In conclusion, our data contribute to the hypothesis that variations in genes encoding for DNA repair proteins increases the susceptibility to MM. These results need to be replicated in larger numbers of patients to confirm and elucidate their role in myeloma pathogenesis.
This research study was supported by Gaziantep University Medical School Research Council. (Project No. TF 09·03).
References
- Kyle RA, Rajkumar SV. Multiple myeloma. NEngl J Med 2004;351:1860–73.
- Rajkumar SV, Kyle RA. Plasma cell disorders. In: , Goldman L, Ausiello D, editors. Cecil textbook of medicine. 23rded. Philadelphia, PA: Saunders; 2007.p. 1426–37.
- Shimizu Y, Kato H, Schull WJ. Studies of the mortality of A-bomb survivors.9. Mortality, 1950–1985: Part 2. Cancer mortality based on the recentlyrevised doses (DS86). Radiat Res 1990;121:120–41.
- Fonscca R, Barlogie B, Bataille R, Bastard C, Bergsagel PL, Chesi M, et al.. Genetics and cytogenetics of multiplmyeloma: a workshop report. Cancer Res 2004;64:1546–58.
- Fenton JA, Pratt G, Rawstron AC, Morgan GJ. Isotype class switching and pathogenesis ofmultiple myelome. Hematol Oncol 2002;20:75–85.
- Bohr VA. DNA repair fine structure and its relationsto genomic instability. Carcinogenesis 1995;16:2885–92.
- Park DJ, Stoehlmacher J, Zhang W, Tsao-Wei DD, Groshen S, Lenz HJ. A Xeroderma pigmentosum group D gene polymorphismpredicts clinical outcome to platinum based chemotherapy in patients withadvanced colorectal cancer. Cancer Res 2001;61:8654–8.
- De Boer J, Hoeijmakers J. Nucleotide excision repair and human syndromes. Carcinogenesis 2000;21:453–60.
- Caldecott KW, Aoufouchi S, Shall S. XRCC1 polypeptide interacts with DNA polymeraseand possibly poly (ADP-riboz) polymerase, and DNA Ligase III is a novel molecular ‘nicksensor’ in vitro. Nucleic Acid Res 1996;24:4387–97.
- Hayden PJ, Tewari P, Morris WD, Staines A, Crowley D, Nieters A, et al.. Variation in NA repair genes XRCC3,XRCC4, XRCC5 and susceptibility to myeloma. Hum MolGenet 2007;16:3117–27.
- Sreeja L, Syamala VS, Syamala V, Hariharan S, Raveendran PB, Vijayalekshmi RV, et al.. Prognostic importance of DNA repairgene olymorphisms of XRCC1 Arg399Gln and XPD Lys751Gln in lung cancer patientsfrom India. J Cancer Res Clin Oncol 2008;134:645–52.
- Chiu CF, Tsai MH, Tseng HC, Wang CL, Wang CH, Wu CN, et al.. A novel single nucleotide polymorphismin XRCC4 gene is associated with oral cancer susceptibility in Taiwanese patients. OralOncol 2008;44:898–902.
- http://ihg2.helmholtz-muenchen.de/cgi-bin/hw/hwa1.pl
- Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. JAm Stat Assoc 1958;53:457–81.
- Cox DR. Regression models and life tables. JR Stat Soc (B) 1972;34:187.
- Durie BGM, Harousseau J-L, Miguel JS, Bladé J, Barlogie B, Anderson K, et al.. International uniform response criteriafor multiple myeloma. Leukemia 2006;20:1467–73.
- Hung RJ, Hall J, Brennan P, Boffetta P. Genetic polymorphisms in the base excisionrepair pathway and cancer risk: a HuGE review. AmJ Epidemiol 2005;126:925–42.
- Rudd MF, Sellick GS, Webb EL, Catovsky D, Houlston RS. Variants in the ATM-BRCA2-CHEK2 axis predisposeto chronic lymphocytic leukemia. Blood 2006;108:638–44.
- Chen S, Tang D, Xue K, Xu L, Ma G, Hsu Y, et al.. DNA repair gene XRCC1 and XPD polymorphismsand risk of lung cancer in a Chinese population. Carcinogenesis 2002;23:1321–5.
- Allan JM, Smith AG, Wheatley K, Hills RK, Travis LB, Hill DA, et al.. Genetic variation in XPD predicts treatmentoutcome and risk of acute myeloid leukemia following chemotherapy. Blood 2004;104:3872–7.
- Mehta PA, Alonzo TA, Gerbing RB, Elliott JS, Wilke TA, Kennedy RJ, et al.. Children’s Oncology Group. XPDLys751Gln polymorphism in the etiology and outcome of childhood acute myeloidleukemia: a Children’s Oncology Group report. Blood 2006;107:39–45.
- Smedby KE, Lindgren CM, Hjalgrim H, Humphreys K, Schöllkopf C, Chang ET, et al.. Variation in DNA repair genes ERCC2,XRCC1 and XRCC3 and risk of follicular lymphoma. CancerEpidemiol Biomarkers Prev 2006;15:258–65.
- Joseph T, Kusumakumary P, Chacko P, Abraham A, Pillai MR. DNA repair gene XRCC1 polymorphisms in childhoodacute lymphoblastic leukemia. Cancer Lett 2005;217:17–24.
- Hill DA, Wang SS, Cerhan JR, Davis S, Davis S, Cozen W, et al.. Risk of non-Hodgkin lymphoma (NHL)in relation to germline variation in DNA repair and related genes. Blood 2006;108:3161–7.
- Shen M, Purdue MP, Kricker A, Lan Q, Grulich AE, Vajdic CM, et al.. Polymorphisms in DNA repair genes andrisk of non-Hodgkin lymphoma in New South Wales, Australia. Haematologica 2007;92:1180–5.
- Matsuo K, Hamajima N, Suzuki R, Andoh M, Nakamura S, Seto M, et al.. Lack of association between DNA baseexcision repair gene XRCC1 Gln399Arg polymorphism and risk of malignant lymphomain Japan. Cancer Genet Cytogenet 2004;140:77–80.
- Chiu CF, Wang CH, Wang CL, Lin CC, Hsu NY, Weng JR, et al.. A novel single nucleotide polymorphismin XRCC4 gene is associated with gastric cancer susceptibility in Taiwan. AnnSurg Oncol 2008;15:514–8.