Abstract
Iron binding protein pirin was isolated as an interactor of the NFIX transcription factor but it can also form complexes with Bcl3 and NF-κB1(p50). Alterations of pirin expression were observed in various tumors and after exposure to pro-carcinogenic oxidative stressors. The aim of the present work was to study the level of pirin transcription in an in vivo model of oxidative stress, namely, in Sod1-deficient mice. We have found that Sod1−/− mice have a significantly elevated level of Pir mRNA in the spleen and kidney but not in the liver, heart, or/and brain. We have also shown that similarly to its human ortholog, the mouse Pir gene transcription level varies significantly between organs. The highest expression was found in the liver and the lowest in the spleen and kidney. Based on literature data, we propose the involvement of Nrf2, AP-1, and NF-κB transcription factors in Pir up-regulation in Sod1−/− mice.
Introduction
Pirin is a nuclear protein that was isolated and characterized in the human as an interactor of the NFIX transcription factor.Citation1 Subsequently, it was revealed that pirin also forms complexes with Bcl3 and NF-κB1 (p50) and therefore, may be implicated in the regulation of the NF-κB-related transcription.Citation2 The crystallographic study of pirin structure showed that the protein binds the iron ion and can be assigned to the functionally diverse cupin superfamily of proteins.Citation3 In addition to its role as a transcription cofactor, pirin also has enzymatic activity and can oxidize the flavonoid quercetin.Citation4 Orthologs of pirin have been found in many organisms ranging from Archaea to mammals. The human PIR gene is located on chromosome X (Xp22.2) and is expressed at different levels in various organs (heart, brain, liver, kidney, lung, pancreas, placenta, skeletal muscle). The highest expression rates were observed in the liver and heart and the lowest in the brain and pancreas.Citation1 Some studies have linked pirin orthologs with apoptosisCitation5,Citation6 and stress responses,Citation7 whereas other have pointed out the deregulation of pirin expression in several human tumors including acute myeloid leukemia,Citation8 melanoma,Citation9,Citation10 and colorectal carcinoma.Citation11 Several studies reported the up-regulation of PIR expression by cigarette smoke in the airway epithelial cells.Citation5,Citation12,Citation13 Hübner et al.Citation12 identified PIR as one of the highly smoking-responsive Nrf2-modulated genes. Nrf2 is an oxidative stress-responsive transcription factor that induces the expression of a battery of detoxifying and antioxidant genes. It binds to specific DNA sequences in the gene's regulatory regions called the antioxidant response elements (AREs).Citation14 Four such AREs have been identified in the PIR promoter but not all of them are thought to be functionally relevant.Citation12 The presence of ARE elements in its promoter and its dependence on the Nrf2 transcription factor strongly suggest that pirin function is related to the cellular response to stress (in particular, oxidative stress). This assumption is supported by papers that reported up-regulation of pirin orthologs in response to various stress conditions in CyanobacteriaCitation7 and to camptothecin-induced lethal damage in the tomato.Citation6
The aim of the present work was to study the level of pirin transcription in an in vivo model of oxidative stress. As mentioned above, there are various studies presenting pirin up-regulation in certain tumors and tumorigenic conditions in specific tissues. Our intention was to perform a more general, whole organism study on the transcription of PIR in different tissues exposed to chronic, pro-carcinogenic oxidative stress conditions. We believe that the transgenic cytosolic superoxide dismutase (Sod1)-deficient mice constitute a suitable model to study transcriptome alterations in such an environment.
Superoxide dismutase (SOD) catalyzes dismutation of the superoxide radical anion leading to hydrogen peroxide and molecular oxygen formation and constitutes a part of the first line of cellular defense against oxidative stress. There are three SOD isoforms in mammals: cytosolic (SOD1, CuZnSOD), mitochondrial (SOD2, MnSOD) and extracellular (SOD3, EC-SOD).Citation15 Sod1−/− mice are characterized by decreased lifespan, elevated incidence of liver cancer,Citation16 and increased sensitivity to pro-oxidative agents (e.g. paraquat and ethanol).Citation17,Citation18 Elevated level of oxidative DNA damage and increased mutation frequency in the liver and kidney of Sod1−/− mice are clear hallmarks of chronic oxidative stress present in these organs.Citation16,Citation19,Citation20 Such stress conditions may trigger activation of several transcription factors, among them Nrf2, AP-1, and NF-κB. Indeed, the increase in NF-κB1 (p50) activity in the kidney of Sod1-deficient mice has recently been reported.Citation20 Despite the lack of information about Nrf2 and AP-1 activity in Sod1−/− mice one can suppose that it is also affected. Because of the postulated pirin involvement in response to stress, its participation in the NF-κB signaling pathway and the presence of ARE elements in its promoter sequence, alterations of pirin expression in Sod1−/− mice are highly probable. In the present work, we compared pirin expression in Sod1-deficient mice and in wild-type mice.
The presence of ARE elements in pirin promoter had been reported only in humans. Since this work was focused on studying the expression of the mouse ortholog of PIR, we show that potential AREs are also present in the sequence of the mouse Pir gene promoter.
Materials and methods
Analysis of the mouse Pir promoter sequence
The sequence upstream of the first exon of the mouse Pir gene (gi149361525:160701363–160707613) was analyzed for the presence of ARE elements using Bioedit software.Citation21
Animals
A breeder pair of mice (strain B6; 129S7-Sod1tm1Leb) heterozygous for a Sod1tm1Leb targeted mutationCitation22 and their progeny were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). Males and females heterozygous for the non-functional Sod1 allele (Sod1+/−) were intercrossed and their progeny were kept at 24–25°C, in 80% humidity with a light–dark cycle of 12 hours. The mice received a standard laboratory diet (Labofeed, Kcynia, Poland) and water ad libitum. Genotyping using DNA isolated from mouse tails was performed by PCR analysis according to the protocol provided by The Jackson Laboratory. Mice homozygous for the non-functional Sod1 allele (Sod1−/−) and control mice homozygous for the wild-type Sod1 allele (Sod1+/+) at the age of 12 months were used in the study. All the experimental procedures involving animals were approved by the local Ethical Commission.
RNA isolation, reverse transcription, and real-time PCR
Liver, kidney, heart, brain, and spleen were excised immediately after death from mice killed by cervical dislocation, flash frozen in liquid nitrogen, and stored at −75°C. Total RNA was extracted from the tissues using the RNeasy Mini Kit (Qiagen, Germany) according to the manufacturer's protocol. One microgram of total RNA was converted to cDNA in a 20 µl reaction volume using High Capacity cDNA Reverse Transcription Kit (Life Technologies, USA) following the manufacturer's instructions. After reaction, cDNA was diluted to 100 µl with de-ionized, nuclease-free H2O. Real-time PCR was performed in 20 µl reaction mixture containing 5 µl of diluted cDNA, 4 µl of de-ionized, nuclease-free H2O, 10 µl of TaqMan Gene Expression Master Mix (Life Technologies), and 1 µl of TaqMan Gene Expression Assay (Life Technologies) for Pir (Assay ID: Mm01721878_m1) or Actb (Assay ID: Mm00607939_s1). All reactions were run in duplicate. PCR amplification was carried out using 7500 Real-Time PCR System (Life Technologies) with an initial 10-minute step at 95°C followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Relative expression levels were calculated using REST 200923 software with Actb as a reference control. Significant differences (P < 0.05) in relative expression were determined by the pair-wise fixed reallocation randomization test built in REST 2009.Citation23 In this test Ct values for reference gene (Actb) and target gene (Pir) were jointly, repeatedly (2000 iterations), and randomly reallocated to control and sample group and the expression ratios were calculated each time. The proportion of expression ratios that were as high as that observed in the experiment gave the P value of the test.
Results
Four putative ARE elements are present in the mouse Pir promoter
The analysis of the sequence 6 kb upstream of the first exon of the mouse Pir gene allowed us to identify four putative ARE elements matching ARE consensus 5′-RTGAYNNNGC-3′. The sequences and positions of the identified elements are presented in .
Table 1. AREs in the promoter sequence of mice Pir gene
Pir expression significantly differs between the organs of wild-type mice
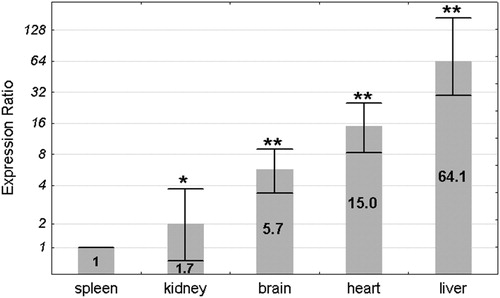
The Pir mRNA levels in spleen, kidney, brain, heart, and liver of 16 (8 males and 8 females) Sod1+/+ mice were estimated by real-time PCR with Actb as reference control. The lowest mean expression was found in the spleen and was set as 1. The expression levels in the other organs were compared to the spleen by the ΔΔCt method and were shown as an expression ratio (fold change) over the spleen (). The highest mean expression was found in the liver (64.1 times higher than in spleen). The mean expression levels in the heart and brain were higher in comparison to the spleen 15 and 5.7 times, respectively. The mean expression in the kidney was only 1.7 times higher in comparison to the spleen. All differences in the mean Pir expression between the organs were statistically significant in a pair-wise fixed reallocation randomization test.Citation23
Figure 1. Mean Pir expression ratio in each organ of the wild-type mice. Expression ratios are presented relative to the spleen, where the expression level was the lowest and for clarity has been set as 1. Values are presented on a logarithmic scale. The exact expression ratio value for each organ is depicted on the respective column. Error bars show standard errors. Asterisks denote the statistical significance of the difference between each organ and the spleen (pair-wise fixed reallocation randomization test; **P < 0.01, *P < 0.05). n = 16 animals (8 males and 8 females).
We also compared the Pir expression in each organ between males and females, but we did not find any gender-related differences (data not shown).
Comparison of Pir expression between Sod1+/+ and Sod1−/− mice
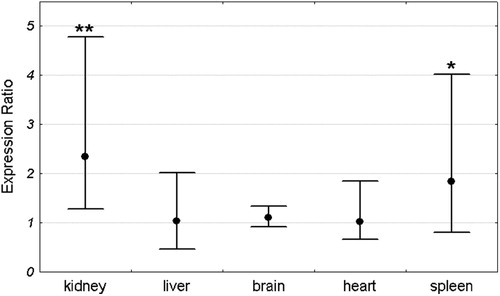
Mean Pir expression in each organ was compared between Sod1+/+ and Sod1−/− mice. For each organ the expression in the Sod1+/+ group was set as 1. The expression level in the Sod1−/− group was compared to the Sod1+/+ by the ΔΔCt method and was shown as an expression ratio (fold change) over the Sod1+/+ group. There were 16 animals in each group (8 males and 8 females). Significant increase in Pir expression in Sod1−/− mice was observed in the kidney and spleen. Mean expression in the kidney of Sod1 deficient mice was 2.54 times higher than in the wild-type animals. Similarly, the mean expression in the spleen of knock-out mice was 2.2 times higher than in the control group. In contrast to the spleen and kidney, there were no differences in Pir expression between Sod1+/+ and Sod1−/− animals in the brain, heart, and liver (). Similar results were obtained when males and females were analyzed separately ( and ).
Figure 2. Pir expression ratio in each organ of Sod1−/− mice relative to wild-type animals. Dots represent mean expression ratios (fold changes) over the Sod1+/+ and error bars represent standard errors. Asterisks denote the statistical significance of the difference between Sod1−/− and Sod1+/+ (pair-wise fixed reallocation randomization test; **P < 0.01). n = 16 animals (8 males and 8 females) with each genotype.
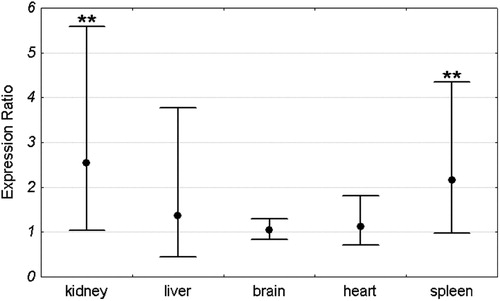
Figure 3. Pir expression ratio in each organ of Sod1−/− male mice relative to wild-type males. Dots represent the mean expression ratios (fold changes) over the Sod1+/+ and error bars represent the standard errors. Asterisks denote the statistical significance of difference between Sod1−/− and Sod1+/+ (pair-wise fixed reallocation randomization test; **P < 0.01, *P < 0.05). n = 8 animals with each genotype.
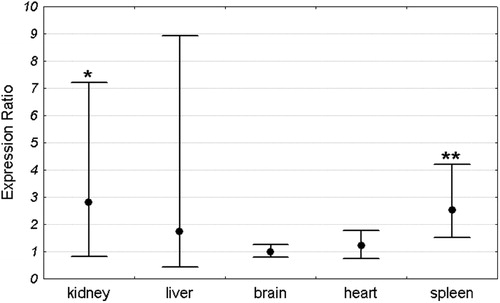
Figure 4. Pir expression ratio in each organ of Sod1−/− female mice relative to wild-type females. Dots represent the mean expression ratios (fold changes) over the Sod1+/+ and error bars represent the standard errors. Asterisks denote the statistical significance of difference between Sod1−/− and Sod1+/+ (pair-wise fixed reallocation randomization test; **P < 0.01, *P < 0.05). n = 8 animals with each genotype.
Discussion
Pirin expression in wild-type mice
The first report concerning pirin expression at mRNA level in different tissues was published by Wendler et al. in 1997.Citation1 It showed that the human PIR gene is differentially expressed in human tissues. Until now, there were no reports referring to the pirin expression pattern in other organisms. In the present paper, we showed that the mouse Pir gene transcription level varies significantly between organs in a way similar to its human ortholog. The highest expression of both mouse and human pirin orthologs was found in the liver. However, whereas Wendler et al. reported that among all human tissues tested the lowest PIR mRNA levels were detected in the brain and pancreas; in our study, the lowest Pir expression was found in the mouse spleen and kidney. Wendler et al. did not study PIR expression in the human spleen and we did not determine Pir expression in the mouse pancreas, so we are not able to compare its expression in these organs between mice and humans. However, the comparison of the pirin transcription in the brain and kidney in both studies brought contradictory results. Whereas in humans the expression in the brain was clearly lower than in the kidney, in mice it was considerably higher, compared to the kidney. This result suggests that the pirin expression pattern can vary between species, but it also could have been an artifact due to different methods of mRNA quantification used in each study (northern blot vs. real-time PCR).
Pirin-coding genes both in mice and in humans are located on the X chromosome. This suggests that their expression may be gender related. Our results did not support this hypothesis, since we found no differences in Pir expression between males and females in any organ studied. This is probably due to the dosage-compensation mechanism, evolved to balance gene expression between males and females that in the case of mammals involve inactivation of one of the two X chromosomes in females.Citation24
Differences in Pir expression between wild-type and Sod1-deficient mice
Previous reports suggesting pirin involvement in stress response and presenting its up-regulation under conditions of oxidative stress caused by cigarette smokeCitation5–Citation7,Citation12,Citation13 encouraged us to test the hypothesis predicting that Pir expression is altered in Sod1-deficient mice. The experimental results supported the hypothesis, since among animals tested, mean Pir expression was significantly higher in the kidney and spleen of the Sod1−/− group, as compared to the Sod1+/+ group. However, the effect of the Sod1 deficiency on Pir expression seems to be organ specific, as it was absent in the liver, heart, and brain. This organ-specific response may be related to differences in the level of Pir expression between organs. It seems meaningful that the effect was observed in the two organs which normally exhibit the lowest Pir expression. It seems plausible to assume that in other organs pirin level is high enough to meet the stress conditions arising from a Sod1 deficiency. It is worth noting that Pir expression in Sod1−/− mice increased to a similar extent both in males and females; considering Pir localization on the X chromosome, this was not an obvious result.
The detailed molecular mechanisms responsible for Pir up-regulation in Sod1−/− mice require further investigation, but the involvement of several transcription factors can be proposed: (1) as shown in this report, putative ARE elements are present not only in the human PIR gene but also in its mouse ortholog. The presence of ARE elements suggests a dependence on Nrf2 transcription factor that is activated in response to oxidative stress and up-regulates the expression of antioxidant genes to restore redox homeostasis. As already mentioned, pirin was identified as one of the Nrf2-modulated genes in response to cigarette smoke;Citation12 (2) Bergman et al.Citation25 reported over-expression of pirin in c-JUN transformed fibroblasts, an observation that strongly suggests the involvement of AP-1 transcription factor in the regulation of pirin expression. In our recent work, we have shown that Fos and Jun (both AP-1 subunits) expression was elevated in the kidney (Fos only) and liver (both Fos and Jun) of Sod1−/− mice.Citation26 It is highly probable that changes in the expression of AP-1 subunits affect its activity, which in turn may have an influence on pirin expression; (3) it is also possible that Pir expression is related to NF-κB activity. It is well established that NF-κB activity is regulated by reactive oxygen speciesCitation27 and increase in NF-κB1 (p50) activity in kidney of Sod1−/− mice has recently been reported.Citation20 Since pirin forms complex with Bcl3 and p50,Citation2 the simultaneous increase in Pir expression and p50 activity in the same organ may not be merely a coincidence. However, we do not know whether it is the increased p50 activity that triggers up-regulation of Pir expression or conversely, whether the elevated pirin level is responsible for the increase in p50 activity. This needs further investigation.
In this work we have proved that the oxidative stress related gene Pir is significantly up-regulated in the mouse kidney and spleen under conditions of permanent oxidative stress resulting from Sod1 deficiency. Various results presenting pirin protein involvement in NF-κB signaling pathway regulation and its dependence on the activation of the stress-responsive transcription factor Nrf2 suggest that this protein may be classified as a putative proto-oncogene whose physiological activity certainly merits further studies.
Acknowledgements
The authors thank Professor Irena Szumiel for critical reading of the paper, Monica Borrin-Flint for the assistance in the preparation of the manuscript and Dr Paweł Lipiński for providing the animal samples gathered during realization of a grant from Ministry of Science and Higher Education 6P04A 00620. This work was supported by a statutory grant for the Institute of Nuclear Chemistry and Technology (to KB, TS, and MK) and the Institute of Agricultural Medicine (to MK).
Conflict of interest statement: None declared.
References
- Wendler WM, Kremmer E, Forster R, Winnacker EL. Identification of pirin, a novel highly conserved nuclear protein. J Biol Chem 1997;272(13):8482–9.
- Dechend R, Hirano F, Lehmann K, Heissmeyer V, Ansieau S, Wulczyn FG, et al. The Bcl-3 oncoprotein acts as a bridging factor between NF-kappaB/Rel and nuclear co-regulators. Oncogene 1999;18(22):3316–23.
- Pang H, Bartlam M, Zeng Q, Miyatake H, Hisano T, Miki K, et al. Crystal structure of human pirin: an iron-binding nuclear protein and transcription cofactor. J Biol Chem 2004;279(2):1491–8.
- Adams M, Jia Z. Structural and biochemical analysis reveal pirins to possess quercetinase activity. J Biol Chem 2005;280(31):28675–82.
- Gelbman BD, Heguy A, O'Connor TP, Zabner J, Crystal RG. Upregulation of pirin expression by chronic cigarette smoking is associated with bronchial epithelial cell apoptosis. Respir Res 2007;8:10.
- Orzaez D, de Jong AJ, Woltering EJ. A tomato homologue of the human protein PIRIN is induced during programmed cell death. Plant Mol Biol 2001;46(4):459–68.
- Hihara Y, Muramatsu M, Nakamura K, Sonoike K. A cyanobacterial gene encoding an ortholog of Pirin is induced under stress conditions. FEBS Lett 2004;574(1–3):101–5.
- Licciulli S, Cambiaghi V, Scafetta G, Gruszka AM, Alcalay M. Pirin downregulation is a feature of AML and leads to impairment of terminal myeloid differentiation. Leukemia 2010;24(2):429–37.
- Licciulli S, Luise C, Zanardi A, Giorgetti L, Viale G, Lanfrancone L, et al. Pirin delocalization in melanoma progression identified by high content immuno-detection based approaches. BMC Cell Biol 2010;11:5.
- Miyazaki I, Simizu S, Okumura H, Takagi S, Osada H. A small-molecule inhibitor shows that pirin regulates migration of melanoma cells. Nat Chem Biol 2010;6(9):667–73.
- Yoshikawa R, Yanagi H, Hashimoto-Tamaoki T, Morinaga T, Nakano Y, Noda M, et al. Gene expression in response to anti-tumour intervention by polysaccharide-K (PSK) in colorectal carcinoma cells. Oncol Rep 2004;12(6):1287–93.
- Hubner RH, Schwartz JD, De BP, Ferris B, Omberg L, Mezey JG, et al. Coordinate control of expression of Nrf2-modulated genes in the human small airway epithelium is highly responsive to cigarette smoking. Mol Med 2009;15(7–8):203–19.
- Spira A, Beane J, Shah V, Liu G, Schembri F, Yang X, et al. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci USA 2004;101(27):10143–8.
- Zhang Q, Pi J, Woods CG, Andersen ME. A systems biology perspective on Nrf2-mediated antioxidant response. Toxicol Appl Pharmacol 2010;244(1):84–97.
- Perry JJ, Shin DS, Getzoff ED, Tainer JA. The structural biochemistry of the superoxide dismutases. Biochim Biophys Acta 2010;1804(2):245–62.
- Elchuri S, Oberley TD, Qi W, Eisenstein RS, Jackson RL, Van RH, et al. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene 2005;24(3):367–80.
- Ho YS, Gargano M, Cao J, Bronson RT, Heimler I, Hutz RJ. Reduced fertility in female mice lacking copper-zinc superoxide dismutase. J Biol Chem 1998;273(13):7765–9.
- Kessova IG, Ho YS, Thung S, Cederbaum AI. Alcohol-induced liver injury in mice lacking Cu, Zn-superoxide dismutase. Hepatology 2003;38(5):1136–45.
- Busuttil RA, Garcia AM, Cabrera C, Rodriguez A, Suh Y, Kim WH, et al. Organ-specific increase in mutation accumulation and apoptosis rate in CuZn-superoxide dismutase-deficient mice. Cancer Res 2005;65(24):11271–5.
- Siomek A, Brzoska K, Sochanowicz B, Gackowski D, Rozalski R, Foksinski M, et al. Cu,Zn-superoxide dismutase deficiency in mice leads to organ-specific increase in oxidatively damaged DNA and NF-kappaB1 protein activity. Acta Biochim Pol 2010;57(4):577–83.
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 1999;41:95–8.
- Matzuk MM, Dionne L, Guo Q, Kumar TR, Lebovitz RM. Ovarian function in superoxide dismutase 1 and 2 knockout mice. Endocrinology 1998;139(9):4008–11.
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 2002;30(9):e36.
- Leeb M, Wutz A. Mechanistic concepts in X inactivation underlying dosage compensation in mammals. Heredity 2010;105(1):64–70.
- Bergman AC, Alaiya AA, Wendler W, Binetruy B, Shoshan M, Sakaguchi K, et al. Protein kinase-dependent overexpression of the nuclear protein pirin in c-JUN and RAS transformed fibroblasts. Cell Mol Life Sci 1999;55(3):467–71.
- Brzóska K, Sochanowicz B, Siomek A, Oliński R, Kruszewski M. Alterations in the expression of genes related to NF-κB signaling in liver and kidney of CuZnSOD-deficient mice. Mol Cell Biochem 2011;353(1–2):151–7.
- Morgan MJ, Liu ZG. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res 2011;21(1):103–15.