Abstract
Objective: Apoptosis is characterized by distinct morphological and biochemical changes that occur upon activation of a family of serine proteases known as caspases. Reactive oxygen species (ROS) induce apoptosis in many cell systems. Nuclear receptor subfamily 4, group A, member 1 (NR4A1) has been shown to induce apoptosis in a number of cell lineages, but can also paradoxically act as a death inhibitory factor. In the current study, we focused on the potential role of NR4A1 in hydrogen peroxide (H2O2)-induced apoptosis of normal human umbilical cord fibroblast (HUC-F2) cells.
Methods: Growth of HUC-F2 cells treated with H2O2 was measured by MTT assay. Analysis of gene expression was performed with a STEP ONE PLUS Real Time PCR system. Inactivation of NR4A1 was treated with siRNA. Apoptosis was measured by Beckman Coulter flow cytometer after inhibition of NR4A1 with siRNA and H2O2 treatment. Caspase -3, -8 and -9 was measured by caspase assay kit.
Results: H2O2 treatment led to enhanced NR4A1 expression. Moreover inhibition of NR4A1 with specific siRNA in HUC-F2 cells triggered an increase in apoptosis and caspase-8 and -3 activities following the addition of H2O2.
Discussion: Our results collectively suggest that NR4A1 is a regulator that inhibits extrinsic apoptosis in HUC-F2 cells during oxidative stress through reduction of caspase-8 and -3 activities.
Keywords:
Introduction
Oxygen is both essential for aerobic life, being the terminal electron acceptor in respiration, and hazardous, because of the potency of reactive oxygen species (ROS). ROS include the superoxide anion (O2−), hydrogen peroxide (H2O2), and singlet oxygen, which are produced by neutrophils and macrophagesCitation1 in the human body. At excess concentrations or in the wrong location, ROS can damage proteins, carbohydrates, polyunsaturated fatty acids, and DNA, and lead to a variety of degenerative processes and diseases.Citation2 Apoptosis, a programmed cell death process characterized by distinct morphological and biochemical changes that take place upon the activation of a family of serine proteases known as caspases,Citation3 plays important roles in the development, homeostasis, and anticancer defence of multicellular organisms.Citation4–Citation6 ROS have been shown to induce apoptosis in several different cell systems.Citation7
Nuclear receptor subfamily 4, group A, member 1 (NR4A1, also known as Nur77 or TR3) is widely expressed but primarily detected in the thymus, osteoblast, liver, and pituitary gland.Citation8,Citation9 NR4A1, a member of an orphan nuclear receptor superfamily, was originally recognized for its role in the regulation of cell survival and differentiation.Citation10,Citation11 Expression of NR4A1 is induced by multiple stimuli in different cell types. NR4A1 has been shown to trigger apoptosis in a number of cell lineages exposed to pro-apoptotic stimuli by directly targeting mitochondria and inducing cytochrome-c release.Citation12–Citation15 Paradoxically, NR4A1 is also reported to act as a death inhibitory factor, blocking cell death induced by ceramideCitation16 or tumour necrosis factor (TNF).Citation17 The finding to date support important roles of these receptors in cell transformation and tumourigenicity via both their anti-apoptotic and pro-apoptotic functions. In particular, the recent identification of a functional ligand (1,1-bis (3′-indolyl)-1-(p-substituted phenyl) methane containing trifluoromethyl, hydrogen, and methoxy substituents) for NR4A1 suggests that members of this superfamily could potentially serve as drug targets for disease indications, such as cancer.Citation18
Although apoptotic and carcinogenic roles of NR4A1 have been discussed with respect to its potential as a therapeutic target in cancer cells, limited reports of NR4A1 activity in normal human cell of human umbilical cord fibroblast (HUC-F2) are documented. Therefore, in the current study, we focused on the mechanisms that determine whether NR4A1 exerts pro- or anti-apoptotic activity in normal human cells. In our experiments, inhibition of NR4A1 with siRNA led to increased apoptosis and caspase-8 activity in H2O2-treated human fibroblast HUC-F2 apoptotic cells. Based on the collective finding, we propose NR4A1 acts as a regulator of HUC-F2 cell death under oxidative stress conditions and prevents extrinsic apoptosis through reducing caspase-8 activity.
Experimental methods
Cell culture
HUC-F2 cells were obtained from Riken BioResource Center Cell Bank (Ibaraki, Japan). Cells were cultured in Alpha-Modified Eagle's Medium (α-MEM) (Cellgro, Manassas, VA, USA) containing 10% foetal bovine serum (FBS) (MP Biomedicals Inc., Solon, OH, USA), 150 IU/ml penicillin (Meiji Seika Kaisha, Tokyo, Japan), and 100 mg/ml streptomycin (Meiji Seika Kaisha) in humidified air at 37°C with 5% CO2.
Cytotoxicity assays
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) colorimetric assay was performed in 96-well plates.Citation19 Briefly, HUC-F2 cells were seeded in 96-well plates at a concentration of 6.0 × 103 cells/well, and incubated at 37°C with 5% CO2. The medium was removed and cells in each well incubated with 50 μl MTT solution (5 mg/ml) for 3 hours at 37°C with 5% CO2. The MTT solution was discarded and 50 μl dimethyl sulphoxide added to dissolve insoluble formazan crystal. Absorbance was measured at 550 nm.
Real-time RT-PCR
Total RNA was extracted from HUC-F2 cells, and cDNA was synthesized as described earlier.Citation20 Briefly, total RNA was extracted using Isogen reagent (Nippon gene, Toyama, Japan), and first-strand cDNA, generated from DNase I-treated total RNA (4 μg) with M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions. Analysis of gene expression was performed with a StepOnePlus Real-Time PCR system (Applied Biosystems Japan, Tokyo, Japan) using SYBR Green PCR Master Mix (Applied Biosystems Japan) and the following primers: NR4A1 sense, 5′-CACAGCTTGCTTGTCGATGTC-3′ and antisense, 5′-ATGCCGGTCGGTGATGAG-3′; NR4A3 sense, 5′-GCATCCCCCATGGTTCAG-3′ and antisense, 5′-TCAGTTGGTGCTCCCCTTGT-3′; β-actin sense, 5′-CTTCTACAATGAGCTGCGTG-3′ and antisense 5′-TCATGAGGTAGTCAGTCAGG-3′; and GAPDH sense 5′-TGCCTCCTGCACCACCAACT-3′ and antisense, 5′-AGTAGAGGCAGGGATGATGT-3′. PCR conditions as follows: first incubation at 95°C for 10 minutes, followed by 40 cycles at 95°C for 15 seconds, and 60°C for 1 minute. Normalization of data was achieved by quantitating the cycle time at an arbitrary fluorescence intensity in the linear exponential phase using StepOnePlus Real-Time system software (Applied Biosystems Japan) by calculating the ratio of the relative concentration of each enzyme relative to that of β-actin and GAPDH cDNA, respectively.
Inactivation of NR4A1 with siRNA
The siRNAs used for the experiment were obtained from Life Technology Japan (Tokyo, Japan). The siRNA sequences for NR4A1 were s6680 (GCAUUAUGGUGUCCGCACAtt), s6679 (CCACUUCUCCACACCUUGAtt), s6678 (GCACCUUCAUGGACGGCUAtt) (Silencer Select Pre-designed siRNA), 232748F11 (CCGGUUCUCUGGAGGUCAUCCGCAA), 232748G01 (GGUCCCUGCACAGCUUGCUUGUCGA), and 232903D12 (CAGCAUUAUGGUGUCCGCACAUGUG) (Stealth RNAi). Negative control siRNAs were used according to the manufacturer's instructions. Lipofectamine 2000 was obtained from Life Technology Japan. For dilution of siRNA, Opti-Modified Eagle's Medium (Opti-MEM) I Reduced Serum Medium (GIBCO-Life Technology Japan) was used. Procedures for inactivation were performed using the manufacturer's protocol. Briefly, Lipofectamine 2000 was diluted with Opti-MEM I Reduced Serum Medium and equilibrated for 5 minutes at room temperature. The siRNA was mixed with Opti-MEM I Reduced Serum Medium, incubated for 20 minutes at room temperature, and added to the culture medium. The final concentrations of siRNA and Lipofectamine 2000 were 10 nM and 0.2%, respectively. HUC-F2 cells were cultured in α-MEM containing 10% FBS and 150 IU/ml to 100μg/ml penicillin to streptomycin in the presence of siRNA and Lipofectamine 2000, followed by incubation in 5% CO2 for 48 hours.
Knockdown efficiency was determined using real-time RT-PCR. To determine off-target effects, we searched for homologous sequences using BLAST.
Apoptosis assay
Cellular apoptosis was detected with flow cytometry (FCM) on a Cell Lab Quanta SC MPL instrument (Beckman Coulter, Tokyo, Japan). Briefly, HUC-F2 cells (8 × 105 cells/ml) were seeded into five wells on a six-well plate and treated with siRNA. After 48 hours, the medium was changed to α-MEM medium containing 200 μM H2O2, followed by incubation for a further 2 hours. Following treatment with H2O2, cells were washed with PBS and stained with Annexin V-FITC (Beckman Coulter), according to the manufacturer's instructions. Control HUC-F2 cells (treated with si-NC or not exposed to H2O2) were subjected to the same protocol.
Caspase assay
Caspase activity was detected with the ApoAlert Caspase Assay Plate (Clontech, CA, USA), which contained fluorogenic substrates specific for caspases-3, -8, and -9 immobilized in the wells of a 96-well plate, according to the manufacturer's instructions. Briefly, pelleted HUC-F2 cells (2 × 105 cells) were incubated with 50 μl of 1× cell lysis buffer on ice for 10 minutes. Cell lysates were centrifuged at 15 000 rpm for 5 minutes at 4°C. Supernatant fractions were transferred to a 96-well plate pre-incubated with 50 μl of 2× reaction buffer/dithiothreitol (DTT) mix at 37°C for 5 minutes. After incubation for 2 hours at 37°C, the release of 7-amino-4-methyl coumarin fluorogenic dye was detected using a microplate reader (excitation: 380 nm, emission: 460 nm) (BioTek Japan, Tokyo, Japan).
Statistical analysis
All data are presented as means ±SEM. Statistical analysis was performed using the StatView (SAS Institute Inc. Cary, NC, USA). P values <0.05 were considered statistically significant, as determined using ANOVA with Fisher's Protected Least Significant Difference (PLSD) post hoc method.
Results
H2O2 induces NR4A1 expression
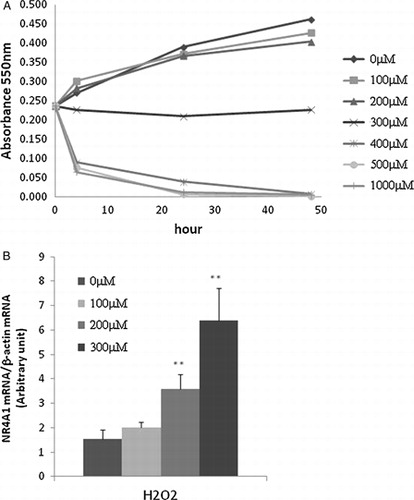
We measured the growth of HUC-F2 cells treated with different concentrations of H2O2 for up to 48 hours with the MTT assay. After 6 hours, cells treated with 400 μM H2O2 or more concentrations died (A). To determine the specific role of NR4A1 in H2O2-mediated apoptosis, its expression patterns under conditions of oxidative stress were measured in cells after 4 hours of treatment using real-time PCR. Our data showed that NR4A1 expression is enhanced by H2O2 in a concentration-dependent manner (B).
Figure 1. Effect of H2O2 on HUC-F2 cells. (A) HUC-F2 cells were treated with 0–1000 μM H2O2 incubated in 5% CO2 for 48 hours, and viability was assessed with the MTT colourimetric assay, as described in Experimental Methods. High concentrations of H2O2 inhibited HUC-F2 cell growth. (B) NR4A1 mRNA levels were determined with real-time PCR in HUC-F2 cells treated with 0–300 μM H2O2 for 4 hours. Results are presented as mean ± standard deviation of three samples in each treatment group. **P < 0.01 versus control. Expression of NR4A1 was increased by H2O2 in a concentration-dependent manner. The differences between the groups were analyzed by ANOVA with Fisher's PLSD post hoc method.
Efficiency of siRNA
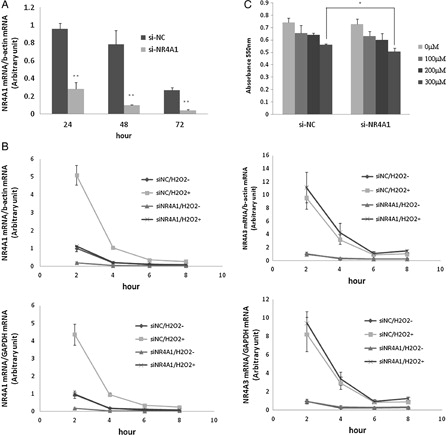
The involvement of NR4A1 in cell death induced by oxidative stress was determined by transient transfection of HUC-F2 cells with NR4A1-specific siRNA (si-NR4A1). NR4A1 expression was significantly reduced upon transfection of cells with NR4A1-targeting siRNA, compared with control siRNA (si-NC), at 24, 48, and 72 hours when the siRNA sequence GGUCCCUGCACAGCUUGCUUGUCGA’ was used (A). The other siRNA examined did not induce >80% inhibition. NR4A1 expression was reduced by 70.4, 87.2, and 83.7%, respectively. Next, HUC-F2 cells were transfected with si-NR4A1 for 48 hours, followed by treatment with 200 μM H2O2 for 2–8 hours. NR4A1 mRNA was rapidly induced in HUC-F2 cells, and the most significant increase in expression was observed at the 2 hours time point (B). To establish whether or not NR4A1 mRNA expression is influenced by H2O2, gene expression profiling was performed with two housekeeping genes (β-actin and GAPDH), NR4A3 expression and additionally monitored to examine off-target effects. The gene expression patterns of NR4A3 were not significantly different. As shown in B, NR4A1 mRNA expression was four-fold higher upon treatment with 200 μM H2O2 for 4 hours, compared with untreated cells. For cells treated si-NC under similar conditions, almost no induction was observed (B). This finding indicates that siRNA of negative control did not have any effect on NR4A1 expression. Si-NR4A1-transfected HUC-F2 cells were subsequently challenged with 0–300 μM H2O2 for 24 hours. Growth of HUC-F2 cells transfected with si-NR4A1 was significantly reduced following exposure to 300 μM H2O2 for 24 hours, as measured with the MTT assay (C).
Figure 2. Effect of si-NR4A1 on mRNA expression and cell growth. (A) HUC-F2 cells were cultured with 10 nM siRNA and 0.2% Lipofectamine 2000 in α-MEM containing 10% fetal bovine serum and 150 IU/ml to 100 μg/ml penicillin to streptomycin medium for 24, 48, and 72 hours in 5% CO2. Real-time PCR analysis showed that NR4A1 expression is reduced by 70.4, 87.2, and 83.7%, respectively. (B) HUC-F2 cells were transfected with si-NR4A1 for 48 hours and treated with 200μM H2O2 for 2–8 hours. Expression levels of NR4A1 and NR4A3 were determined using real-time PCR. Normalization of data was achieved by quantitating the cycle time at an arbitrary fluorescence intensity in the linear exponential phase using StepOnePlus Real-Time system software by calculating the ratio of the concentration of each enzyme relative to that of β-actin and GAPDH cDNA, respectively. (C) Effects of the H2O2 concentration on the HUC-F2 cells treated with si-NR4A1 were determined with the MTT assay. Specifically, HUC-F2 cells were transfected with si-NR4A1 or si-NC, incubated in 5% CO2 for 48 hours, and treated with H2O2. After 24 hours, cell viability was assessed with the MTT assay. Results are presented as mean ± standard deviation of three samples in each treatment group. The differences between the groups were analyzed by ANOVA with Fisher's PLSD post hoc method. *P < 0.05 versus control; **P < 0.01 versus control.
Detection of apoptosis
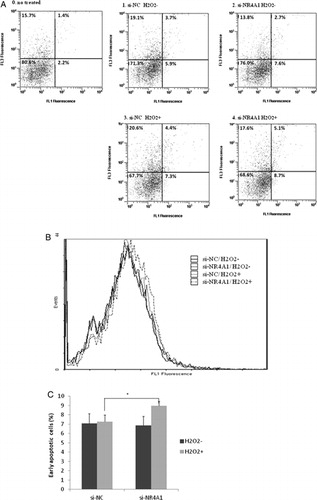
To compare the proportion of early apoptotic cells, HUC-F2 cells were transfected with siRNA and treated with 200 μM H2O2 at concentrations below that leading to reduced cell viability. Apoptosis was determined using FCM. HUC-F2 cells were transfected with si-NR4A1 for 48 hours, treated with 200 μM H2O2 for 2 hours and measured with FCM (A4). Cells transfected with si-NC without H2O2 (A1), si-NR4A1 without H2O2 (A2), and si-NC with 200 μM H2O2 (A3) were additionally analyzed using FCM (A). The peak of the annexin V-FITC intensity histogram for cells treated with si-NR4A1 and H2O2 was shifted rightward, compared with the control (B). The percentages of early apoptotic cells (bottom right) were additionally compared among the four samples. FCM analysis showed an increase in early apoptotic HUC-F2 cells transfected with si-NR4A1 and treated with H2O2 to 9.0% from 7.3% (si-NC with H2O2, P = 0.013) (C), suggesting a prosurvival or anti-apoptotic function of NR4A1.
Figure 3. Determination of cell apoptosis using FCM. HUC-F2 cells transfected with si-NC or si-NR4A1 were left untreated or treated with H2O2 at a concentration of 200 μM for 2 hours, processed for annexin V/propidium iodide (PI) staining, and analyzed using Beckman Coulter FCM. (A) The apoptotic region was determined in relation to normal untreated HUC-F2 cells. Viable cells are located in the bottom left corner (negative for annexin V-FITC and PI), as observed for most normal HUC-F2 cells (A0). Early apoptotic cells are present in the bottom right corner (annexin V-FITC positive). Late apoptotic cells showing signs of progressive cellular membrane and nuclear damage are in the upper right corner (double positive), and the four divisions were fixed with this gating. Cells transfected with si-NC without H2O2 (A1), si-NR4A1 without H2O2 (A2), si-NC with 200 μM H2O2 (A3), and si-NR4A1 with 200 μM H2O2 (A4) were analyzed. (B) Distribution of annexin V-positive cells. Annexin V-FITC intensity histograms of the four samples. The vertical axis shows the intensity of annexin V-FITC and the horizontal axis depicts cell number. (C) Percentage of early apoptotic cells in the four groups. Results are presented as mean ± standard deviation of five samples in each treatment group. The differences between the groups were analyzed by ANOVA with Fisher's PLSD post hoc method.*P < 0.05 versus control. FL1, annexin V-FITC; FL3, PI.
Activation of caspase-3, -8, and -9
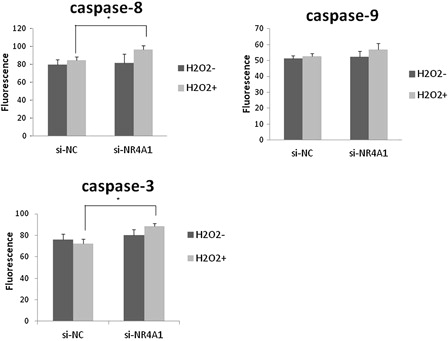
Caspase activation plays a central role in the execution of apoptosis. The key components of the biochemical pathways of caspase activation have been identified.Citation21 Two pathways of caspase activation have been extensively characterized to date, specifically, cell surface death receptor and the mitochondria-initiated pathways. In the cell surface death receptor pathway, activation of caspase-8 following its recruitment to the death-inducing signalling complex is the critical event that transmits the death signal. In the mitochondria-initiated pathway, caspase activation is triggered by the formation of a multimeric Apaf-1/cytochrome-c complex that is fully functional in recruiting and activating procaspase-9. Activated caspase-8 and -9 cleave and activate downstream caspases-3, -6, and -7. In our experiments, activities of caspase-3 and -8 of NR4A1 knockdown cells treated with 200 μM H2O2 were significantly higher than those of negative control cells. Caspase-9 activity was also elevated, but not to a significant extent (Fig. 4).
Figure 4. Assay for caspase-3, -8, and -9 activities. HUC-F2 cells transfected with si-NC or si-NR4A1 were treated without or with H2O2 at a concentration of 200 μM for 2 hours. Caspase activity was measured using ApoAlert Caspase Assay plates. Each group contained four dishes. The caspase activity of each dish was measured in duplicate, and the average of the two measurements used. Results are presented as mean ± standard deviation of four dishes in each treatment group. The differences between the groups were analyzed by ANOVA with Fisher's PLSD post hoc method. *P < 0.05 versus control.
Discussion and conclusions
NR4A1 has been shown to induce apoptosis in a number of cell lineages exposed to pro-apoptotic stimuli by directly targeting mitochondria and promoting cytochrome-c release.Citation12–Citation15 Paradoxically, NR4A1 can act as a death inhibitory factor, blocking cell death induced by ceramideCitation16 and TNF.Citation17 In the current study, HUC-F2 cells treated with H2O2 at concentrations of 400 μM H2O2 or higher died, and growth stopped at 300 μM H2O2. Expression of NR4A1 was induced by H2O2 in a concentration-dependent manner suggesting that NR4A1 plays a role in HUC-F2 cell death induction under oxidative stress conditions ().
NR4A1 silencing mediated via siRNA has led to a drastic reduction in cell growth/survival in many cancer cell lines.Citation18 This reduced growth correlates well with increased levels of apoptosis. In this study, growth of si-NR4A1-transfected HUC-F2 cells treated with H2O2 was significantly reduced (). FCM analysis also showed an increased proportion of early apoptotic cells in HUC-F2 transfected with si-NR4A1 and treated with H2O2 (). The collective results clearly support a prosurvival or anti-apoptotic function of NR4A1.
Various apoptotic pathways exist that can be distinguished based on the adapters and initiator caspases involved. Most apoptotic programmes fall into either the extrinsic or intrinsic category. Extrinsic apoptosis is triggered by extracellular cues delivered in the form of ligands binding to death receptors (DR). The binding of a ligand to its DR recruits monomeric procaspase-8 protein to be recruited to the death-inducing signalling complex.Citation22 Intrinsic apoptosis is also known as mitochondrial apoptosis since it depends on factors released from mitochondria. This pathway is activated by many cellular stress factors, including growth factor deprivation.Citation23 The initiator caspase responsible for the intrinsic apoptosis pathway is caspase-9, which is activated by dimerization induced when the caspase-9 CARD domain binds to the adapter protein apoptotic protease-activating factor-1.Citation24 Activated caspase-8 and -9 cleave and activate downstream caspases, such as caspases-3, -6, and -7.Citation25 In this study, caspase-3 and -8 activities were elevated in cells transfected with si-NR4A1 and exposed to H2O2, while that of caspase-9 activity was not significantly affected (Fig. 4). The data suggest that NR4A1 inhibits extrinsic apoptosis by reducing caspase-8 and caspase-3 activities.
NR4A1 has been previously implicated in cell growth/survival and apoptosis. In this study, we have demonstrated that NR4A1 inhibits apoptosis caused by oxidative stress in HUC-F2 cells. However, even upon NR4A1 knockdown, many of the cells survived. The most likely explanation for this finding is that NR4A2 and NR4A3 compensate for NR4A1 function, since all three orphan receptors bind the same responsive element of AAAGGTCA.Citation18
Interestingly, NR4A1 expression varies in diverse cell types after stimulations with different reagents. NR4A1 translocation is reported to be involved in myocardial ischemia/reperfusion-induced apoptotic injury of cardiac myocytes subjected to metabolic syndrome.Citation26 It would be interesting to determine whether H2O2 treatment similarly causes cytoplasmic shuttling of NR4A1 in normal fibroblasts. Furthermore, to understand the physiological role of NR4A1 in normal fibroblasts, it is necessary to ascertain whether NR4A1-mediated inhibition of extrinsic apoptosis is specific to HUC-F2 cells or common in normal fibroblasts. Nevertheless, in the current study, we showed that NR4A1 acts as a regulator that inhibits extrinsic apoptosis in HUC-F2 cells during oxidative stress by reducing caspase-8 and -3 activities. Since NR4A1 plays an essential role in DNA repair,Citation27 we are further interested in determining whether inhibition of NR4A1 causes more DNA damage in normal fibroblasts which will be the focus of our future research.
Disclaimer statements
Contributors Yuri Shimizu and Riko Miyakura conducted the experiments and Yuzuru Otsuka planned this research.
Funding This work was supported by Japan Society for the Promotion of Science fund.
Conflicts of interest None.
Ethics approval Ethics approval and consent was not needed.
Acknowledgments
This work was supported by a Grant-in-Aid (no. 25282020) from the Japanese Ministry of Education, Science, Sports, and Culture, Japan.
References
- Fantone JC, Ward PA. Role of oxygen-derived free radicals and metabolites in leukocyte-dependent inflammatory reactions. Am J Pathol 1982;107(3):395–418.
- Alho H, Leinonen J. Total antioxidant activity measured by chemiluminescence methods. Methods Enzymol 1999;299:3–15.
- Nicholson DW. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ 1999;6(11):1028–42.
- Ivanov VN, Bhoumik A, Ronai Z. Death receptors and melanoma resistance to apoptosis. Oncogene 2003;22(20):3152–61.
- Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell 1997;88(3):347–54.
- Rudin CM, Thompson CB. Apoptosis and disease: regulation and clinical relevance of programmed cell death. Annu Rev Med 1997;48:267–81.
- Simon HU, Haj-Yehia A, Levi-Schaffer F. Role of reactive oxygen species (ROS) in apoptosis induction. Apoptosis 2000;5(5):415–8.
- Zetterström RH, Solomin L, Mitsiadis T, Olson L, Perlmann T. Retinoid X receptor heterodimerization and developmental expression distinguish the orphan nuclear receptors NGFI-B, Nurr1, and Nor1. Mol Endocrinol 1996;10(12):1656–66.
- Hsu HC, Zhou T, Mountz JD. Nur77 family of nuclear hormone receptors. Curr Drug Targets Inflamm Allergy 2004;3(4):413–23.
- Liang B, Song X, Liu G, Li R, Xie J, Xiao L, et al. Involvement of TR3/Nur77 translocation to the endoplasmic reticulum in ER stress-induced apoptosis. Exp Cell Res 2007;313(13):2833–44
- Moll UM, Marchenko N, Zhang XK. p53 and Nur77/TR3– transcription factors that directly target mitochondria for cell death induction. Oncogene 2006;25(34):4725–43.
- Cheng Z, Völkers M, Din S, et al. Mitochondrial translocation of Nur77 mediates cardiomyocyte apoptosis. Eur Heart J 2011;32(17):2179–88.
- Chen HZ, Zhao BX, Zhao WX, Li L, Zhang B, Wu Q. Akt phosphorylates the TR3 orphan receptor and blocks its targeting to the mitochondria. Carcinogenesis 2008;29(11):2078–88.
- Zhan YY, Wu Q. Translocation of orphan receptor TR3 from nuclei to mitochondria induced by staurosporine. Ai Zheng 2004;23(12):1593–8.
- Yu H, Kumar SM, Fang D, Acs G, Xu X. Nuclear orphan receptor TR3/Nur77 mediates melanoma cell apoptosis. Cancer Biol Ther 2007;6(3):405–12.
- Brás A, Albar JP, Leonardo E, de Buitrago GG, Martínez-A C. Ceramide-induced cell death is independent of the Fas/Fas ligand pathway and is prevented by Nur77 overexpression in A20 B cells. Cell Death Differ 2000;7(3):262–71.
- Suzuki S, Suzuki N, Mirtsos C, et al. Nur77 as a survival factor in tumor necrosis factor signaling. Proc Natl Acad Sci USA 2003;100(14):8276–80.
- Li QX, Ke N, Sundaram R, Wong-Staal F. NR4A1, 2, 3--an orphan nuclear hormone receptor family involved in cell apoptosis and carcinogenesis. Histol Histopathol 2006;21(5):533–40.
- Liu WK, Ho JC, Qin G, Che CT. Jolkinolide B induces neuroendocrine differentiation of human prostate LNCaP cancer cell line. Biochem Pharmacol 2002;63(5):951–7.
- Ueta E, Suzuki E, Nanba E, Tadokoro Y, Otsuka Y, Kurata T. Regulation of cigarette smoke-induced cytochrome P4501A1 gene expression in osteogenic disorder Shionogi rat liver and in lung by large ascorbic acid dose. Biosci Biotechnol Biochem 2001;65(11):2548–51.
- Anilkumar TV, Sarraf CE, Alison MR. The biology and pathology of programmed cell death (apoptosis). Vet Hum Toxicol 1992;34(3):251–4.
- Kaufmann T, Strasser A, Jost PJ. Fas death receptor signalling: roles of Bid and XIAP. Cell Death Differ 2012;19(1):42–50.
- Brenner D, Mak TW. Mitochondrial cell death effectors. Curr Opin Cell Biol 2009;21(6):871–7.
- Shiozaki EN, Chai J, Shi Y. Oligomerization and activation of caspase-9, induced by Apaf-1 CARD. Proc Natl Acad Sci USA 2002;99(7):4197–202.
- Kruidering M, Evan GI. Caspase-8 in apoptosis: the beginning of ‘the end’? IUBMB Life 2000;50(2):85–90.
- Xu A, Liu J, Liu P, Jia M, Wang H, Tao L. Mitochondrial translocation of Nur77 induced by ROS contributed to cardiomyocyte apoptosis in metabolic syndrome. Biochem Biophys Res Commun 2014;446(4):1184–9.
- Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal 2006;4:e002.
