Abstract
Background
Cytogenetically normal acute myeloid leukemia (AML) represents nearly half of newly diagnosed de novo AML cases. XPD is one of the DNA repair proteins, whose genetic polymorphisms are thought to affect their function as regards response to chemotherapeutic drugs and chemotherapy-induced toxicities.
Subjects and methods
We investigated the XPD Asp312Asn and Lys751Gln polymorphisms by polymerase chain reaction-restriction fragment length polymorphism in 51 newly diagnosed cytogenetically normal de novo AML patients. The response to the standard induction chemotherapy protocol and chemotherapy-induced toxicities were monitored.
Results
The XPD Asp312Asn GG genotype was the most frequent (57%) followed by the GA variant (37%), and the AA variant was the least frequent (6%). As regards the XPD Lys751Gln polymorphism, the AA genotype was the most frequent (49%), followed by the AC (39%) and CC (12%) variants. These variants were not associated with age, sex, FAB subtype, CNS infiltration, chemotherapy-induced hepatotoxicity, nephrotoxicity, or metabolic toxicity. The XPD Lys751Gln CC polymorphic variant was associated with chemotherapy-induced cardiotoxicity and lower chance to achieve response to induction chemotherapy.
Conclusion
XPD Lys751Gln and not Asp312Asn polymorphism was associated with chemotherapy-induced cardiotoxicity and response to induction chemotherapy in newly diagnosed cytogenetically normal AML patients. Pretreatment assay of XPD Lys751Gln may help to anticipate cardiotoxicity in those at risk. Moreover, it may be considered a prognostic marker in AML cases. However, further large scale research is needed to verify its usefulness.
Keywords:
Introduction
Acute myeloid leukemia (AML) is a clonal, malignant disease of hematopoietic tissues that is characterized by accumulation of abnormal (leukemic) blast cells, principally in the marrow, and impaired production of normal blood cells. Thus, the leukemic cell infiltration in marrow is accompanied, nearly invariably, by anemia and thrombocytopenia.Citation1 The overall annual incidence of AML is 3.7 per 100 000 persons, and age-dependent mortality is 2.7 to nearly 18 per 100 000 persons.Citation2 In adults, AML accounts for 80–90% of cases of acute leukemia.Citation3
There is considerable heterogeneity between cases of AML as regards morphology, immunological phenotype, associated cytogenetic and molecular abnormalities, and patterns of gene expression.Citation4 A number of clinical and biologic features predict prognosis in AML including clinical state, morphology, surface/enzyme markers, cytogenetics, and molecular markers.Citation3
The backbone of AML treatment for 30 years has been the combination of daunorubicin and cytarabine for remission induction chemotherapy protocol.Citation4 These chemotherapeutic drugs bring about their effect by causing DNA damage.Citation5 This damage causes activation of DNA-repair mechanisms in the different phases of the cell cycle. Cells resume cell-cycle progression once damage has been repaired, whereas cells which fail to repair the DNA damage undergo permanent cell-cycle arrest or apoptosis.Citation6
There are six pathways of DNA repair: homologous recombination repair, non-homologous end-joining, base excision repair, mismatch repair, nucleotide excision repair (NER), and methyltransferase repair.Citation7 NER is a major DNA repair pathway in eukaryotic cells. It involves more than 30 proteins including Xeroderma pigmentosum (XP) groups A to G.Citation8
Xeroderma pigmentosum complementation group D (XPD) gene encodes a helicase which is a component of transcription factor IIH (TFIIH) and functions in transcription initiation and NER in eukaryotes.Citation9 Functional DNA repair capacity is thought to differ significantly between the different polymorphic variants of XPD.Citation10,Citation11
Among the polymorphic variants described for XPD gene are Lys751Gln (A > C) and Asp312Asn (G > A).Citation12 The association between these variants and the response to chemotherapy in AML has been subjected to recent research in order to predict the prognosis of AML patients.Citation5,Citation13
In this work, we aimed to investigate the association of XPD polymorphism with the response to remission induction chemotherapy. In addition, we explored the association of XPD polymorphism with chemotherapy-induced toxicities during remission induction in patients with de novo cytogenetically normal AML.
Subjects and methods
This study included 51 patients with newly diagnosed de novo cytogenetically normal AML presented to the Department of Hematology, Medical Research Institute, Alexandria University. Patients over 60 years old and those with co-morbid conditions (liver, kidney, or heart disease) were excluded from the study. All patients were subjected at presentation to full medical history taking, thorough clinical examination, routine laboratory, and imaging workups.
Patients received the standard remission induction protocol (3 + 7) comprising: Cytosine Arabinoside 100 mg/m2 on days 1–7 by continuous intravenous infusion and daunorubicin 45 mg/m2 on days 1–3 by intravenous push.Citation4,Citation14 Patients were monitored for toxic effects induced by chemotherapy, namely gastrointestinal toxicity (nausea, diarrhea or mucositis), hepatotoxicity (increased serum bilirubin, alkaline phosphatase, or transaminases levels), nephrotoxicity (increased serum creatinine or presence of proteinuria), metabolic toxicity (hypoglycemia, hyperglycemia, hypocalcemia, hypokalemia, hyponatremia, or hypomagnesemia) and cardiac toxicity (conduction abnormalities, palpitations, prolonged QTc interval, arrhythmias, ischemia/infarction, hypertension, hypotension, left or right ventricular dysfunction, pericarditis, pulmonary hypertension, or cardiomyopathy). Monitoring included clinical examination, laboratory evaluation (liver and kidney functions, blood electrolytes, and cardiac enzymes assays), electrocardiography, and imaging studies including echocardiography. The grade of toxicity was recorded according to the National Cancer Institute common terminology criteria (version 4.0).Citation15
Response to chemotherapy was evaluated after regeneration from chemotherapy-induced bone marrow aplasia.Citation4 Patients who failed to achieve complete remission were subjected to a second similar course of chemotherapy.
Molecular studies for XPD polymorphisms
Sample collection and DNA extraction
Three milliliters of venous blood were collected before receiving chemotherapy after obtaining signed consent. Immediately after collection, whole blood was stored in aliquots at −20 °C until assayed. Genomic DNA was extracted from leukocytes using Gene JET DNA purification kit (Feremntas, Germany) according to the manufacturer's instructions.
Polymerase chain reaction-restriction fragment length polymorphism assay for XPD Lys751Gln (A > C)
The XPD Lys751Gln (A > C) genotypes were determined by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis of DNA samples collected previously.Citation16 The PCR primers were: forward, 5′-GCCCGCTCTGGATTATACG-3′; and reverse, 5′-CTATCATCTCCTGGCCCCC-3′. PCR was performed in 50-μl containing 2 mM MgCl2, 0.04 mM deoxynucleotide triphosphates, 2.5 units of Taq polymerase, and the manufacturer's buffer [20 mM Tris-HCI (pH 8.4) and 50 mM KCl]. After an initial denaturation at 94°C for 3 minutes, there were 38 cycles of 45 seconds at 94°C, 45 seconds at 60°C, and 60 seconds at 72°C, and then a final extension step of 7 minutes at 72°C. After overnight digestion of the PCR product with PstI, 15 µl of the digested products were resolved on a 3% agarose gel containing ethidium bromide. The homozygous wild-type allele (Lys 751) produced two DNA bands (290 and 146 bp), whereas the mutant allele (Gln 751) produced three DNA bands (227, 146, and 63 bp). Heterozygotes displayed all four bands (290, 227, 146, and 63 bp).
PCR-RFLP assay for XPD Asp312Asn (G > A)
XPD Asp312Asn (G > A) polymorphism was analyzed by PCR-RFLP method.Citation16 The oligonucleotide primers used were: 5′-CTGTTGGTGGGTGCCCGTATCTGTT-GGTCT-3′ and 5′-TAATATCGGGG CTCACCCTGCAGCACTTCCT-3′. PCR was performed in 25 µl reaction mixtures containing 1.5 mM MgCl2, 0.2 mM deoxynucleotide triphosphates, 3% DMSO, 0.2 µM primers, 1 µg of template DNA, and 1.5 units of Taq polymerase in PCR buffer (10 mM Tris-HCl (pH 9.0 at 25°C), 50 mM KCl, and 0.1% Triton X-100 (Promega)). After an initial denaturation at 94°C for 4 minutes, the DNA was amplified by 30 cycles of 30 seconds at 94°C, 30 seconds at 60°C, and 60 seconds at 72°C, and then by a final extension step of 5 minutes at 72°C. Fifteen microliters of the PCR product was digested with StyI for 8 hours at 37°C. The digestion products were then resolved on a 3% agarose gel (5 V/cm) containing ethidium bromide. The homozygous wild type (Asp/Asp) was identified by two DNA bands (507 and 244 bp), the homozygous mutant type (Asn/Asn) produced three bands (474, 244, and 33 bp); and heterozygotes (Asp/Asn) displayed all four bands (507, 474, 244, and 33 bp).
Statistical analysis of the data
Data were fed to the computer using IBM SPSS software package, version 20.0. Comparison between different groups regarding categorical variables was tested using χ2 test. When more than 20% of the cells have expected count <5, correction for χ2 was conducted using Monte Carlo correction. Parametric tests were used for normally distributed data, while non-parametric tests were used for abnormally distributed data. Significance test results are quoted as two-tailed probabilities. Significance of the obtained results was judged at the 5% level.
Results
Fifty one newly diagnosed de novo cytogenetically normal AML patients were enrolled in this study (28 males and 23 females). Age of patients ranged from 19 to 57 years with a median of 33 years. AML FAB subtype M2 was the most frequent while FAB subtype M7 was the least frequent among the studied patients. Six patients had CNS infiltration (11.8%). Among the chemotherapy-induced toxicities, hepatotoxicity was the most frequent (27%) followed by metabolic toxicity (24%), while nephrotoxicity and cardiac toxicity were the least frequent (13%) ().
Table 1. XPD Asp312Asn and Lys751Gln polymorphisms in relation to the studied clinical and laboratory parameters
Thirty patients (59%) achieved complete response while 21 patients (41%) failed to respond to induction chemotherapy. Among the responders, 19 patients achieved the complete response after one course of induction chemotherapy while 11 patients achieved it after 2 courses ().
The XPD Asp312Asn GG genotype was the most frequent (57%) followed by the GA variant (37%), and the AA variant was the least frequent (6%) (). No significant association was found between these variants and any of the studied parameters, namely age, sex, FAB subtypes, CNS infiltration, and different types of chemotherapy-induced toxicities ().
Figure 1. PCR products (A) and digestion products (B) for Asp312Asn polymorphism of 6 cases. The first lane to the left is DNA ladder (50–1000 base). Lanes 2, 4, 5, and 6 represent GA genotype. Lanes 3 and 7 represent GG genotype.
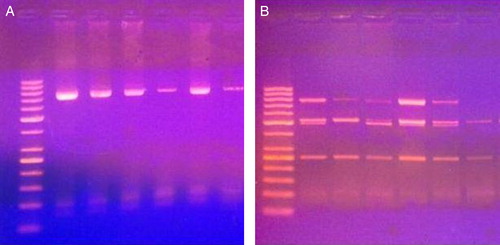
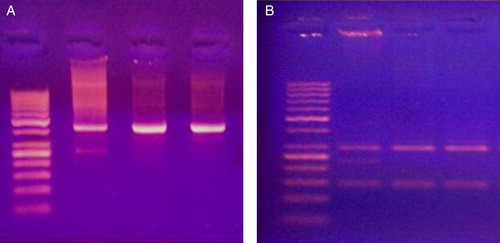
As regards the XPD Lys751Gln polymorphism, the AA genotype was the most frequent (49%), followed by the AC (39%) and CC (12%) variants () (). These variants were not associated with age, sex, FAB subtype, CNS infiltration, chemotherapy-induced hepatotoxicity, nephrotoxicity, or metabolic toxicity. However, while they were significantly associated with chemotherapy-induced cardiotoxicity, which was more frequent among patients having the CC polymorphic variant. In addition, the response to induction chemotherapy was significantly associated with these polymorphic variants with more patients having the AA genotype achieved complete remission ().
Figure 2. PCR products (A) and digestion products (B) for XPD Lys751Gln polymorphism of 3 cases. The first lane to the left is DNA ladder (50–1000 base). Lane 2 represents AC genotype. Lanes 3 and 4 represent AA genotype.
The response to induction chemotherapy among the studied patients was not associated with the age, sex, FAB subtype, or XPD Asp312Asn polymorphism, while it was significantly associated with CNS infiltration and XPD Lys751Gln polymorphism. No patient with CNS infiltration achieved complete response and patients with the C allele of XPD Lys751Gln polymorphism had lower chance to achieve complete response ().
Table 2. The response of acute myeloid leukemia patients to induction chemotherapy in relation to the studied parameters
Discussion
AML is a highly malignant and cytogenetically heterogeneous type of cancer. Cytogenetically normal AML accounts for approximately 40–50% of adult patients with AML and represents a heterogeneous group with an intermediate prognosis.Citation17
Human DNA repair mechanisms protect the genome from DNA damage caused by endogenous and exogenous agents including chemotherapy.Citation18 XPD gene product is a helicase that is a component of the TFIIH complex. XPD plays a role in transcription and NER through reversal of DNA crosslinks and oxidative damage induced by chemotherapy. XPD polymorphic variants are supposed to possess different DNA repair capacities and consequently, result in variable effects of the chemotherapeutic agents.Citation18,Citation19
In this study, the XPD Asp312Asn polymorphism was not associated with chemotherapy-induced toxicities or with the response to induction chemotherapy. As regards the XPD Lys751Gln polymorphism, it was not associated with age, sex, FAB subtype, CNS infiltration, chemotherapy-induced hepatotoxicity, nephrotoxicity, or metabolic toxicity. However, patients with CC genotype (Gln/Gln) had a higher chance to suffer chemotherapy-induced cardiotoxicity.
According to Shen and his colleagues, XPD Lys751Gln is the main polymorphism that induces amino acid changes in the protein and consequently, modifies its DNA repair ability.Citation20 As described by Spitz et al.Citation21, Rzeszowska-Wolny et al.,Citation22 and Shen et al.,Citation23 variant allele of XPD gene codon 751 (C allele) is associated with impaired DNA repair activity. The impaired DNA repair activity was reported to enhance drug toxicities.Citation13,Citation24 On the contrary, a report by Lunn et al.Citation25 relates suboptimal repair of X-ray-induced DNA damage to the common allele (A). However, this can be attributed to the diferenct genotoxic element evaluated and to the in vitro model of their study.
It has been generally agreed with that toxicity from chemotherapeutic drugs depends on the pharmacological effects and drug dosages of regimen, patient's performance status, organ functions, and previous treatments. However, the toxicity may be quite different even though the patients with the similar body status received the same drug dosage. It is now hypothesized that the toxicity could vary for patients with different genotypes; a milestone towards the individualized medicine.
The reason for which, cardiotoxicity was significantly associated with the XPD Lys751Gln polymorphism is not clear. However, recent attention was focused on the association between chemotherapy-induced cardiotoxicity and DNA polymorphisms in order to help selecting the chemotherapeutic regimen most likely to benefit each individual patient.Citation26
A report by Guven et al.Citation27 studied the XPD Lys751Gln polymorphism in patients with coronary artery disease. They found an association between the variant allele C and the evidence of genotoxicity in their patients. It has been suggested that oxidative stress and the generation of reactive oxygen species may play an important role in the induction of DNA damage.Citation28 NER together with base excision repair is responsible for repair of the oxidative DNA damage induced by chemotherapy.Citation29 Accordingly, genetic polymorphisms in the XPD DNA repair gene may influence individual variation in DNA repair capacity in response to chemotherapy, leading to increased risk of developing cardiotoxicity in those having the inefficient C allele.
In this work, an association between the response to chemotherapy and the XPD Lys751Lys common genotype (AA) was found. This issue was raised by different authors. However, none of the them reported similar findings. Kuptsova-Clarkson et al.Citation13, Spitz et al.,Citation21 and Shen et al.Citation23 found the variant allele of XPD gene codon 751 (C allele) to be associated with impaired DNA repair activity. They concluded that impaired DNA repair activity due to this variant, led to lower resistance to chemotherapy and higher anticipation to achieve complete remission (CR) and longer survival. On the other side, Qiu et al.Citation30 and Rumiato et al.Citation31 denied any association between this polymorphism and response to chemotherapy.
Up to our best knowledge, no reports studied this polymorphism in newly diagnosed cytogenetically normal AML patients. A possible explanation for our finding may be the intact DNA repair machinery granted by the common (A) allele limits the genome mutations, which would cause drug resistance, within the leukemic clone. Consequently, a better response to chemotherapy is achieved. Obviously, further research is required before establishing a firm conclusion in this regard.
Conclusion
XPD Lys751Gln and not Asp312Asn polymorphism was associated with chemotherapy-induced cardiotoxicity and response to induction chemotherapy in newly diagnosed cytogenetically-normal AML patients. Pretreatment assay of XPD Lys751Gln might help to anticipate cardiotoxicity in those at risk. Moreover, it may be considered a prognostic marker in AML cases. However, further large scale research in needed to verify its usefulness.
Acknowledgement
We here appreciate the support of the Hematology Department, Medical Research Institute, Alexandria University.
References
- Liesveld JL, Lichtman MA. Acute myelogenous leukemia. In: Kaushansky K, Lichtman MA, Beutler E, Kipps TJ, Seligsohn U, Prchal JT, (eds.) Williams Hematology. 8th ed. New York: McGraw-Hill; 2010. pp. 1277–330.
- Deschler B, Lubbert M. Acute myeloid leukemia: epidemiology and etiology. Cancer 2006;107:2099–107.
- Baer MR, Greer JP. Acute myeloid leukemia in adults. In: Greer JP, Foerster J, Rodgers GM, Paraskevas F, Glader B, Arber DA, Means RT, (eds.) Wintrobe's clinical haematology. 12th ed. Philadelphia: Lippincott Williams & Wilkins; 2009. pp. 1843–89.
- Burnett AK, Venditti A. Acute myeloid leukaemia. In: Hoffbrand AV, Catovsky D, Tuddenham EG, Green AR, (eds.) Postgraduate haematology. 6th ed. Oxford: Blackwell Publishing; 2010. pp. 415–32.
- Kuptsova N, Kopecky KJ, Godwin J, Anderson J, Hoque A, Willman CL, et al. Polymorphisms in DNA repair genes and therapeutic outcomes of AML patients from SWOG clinical trials. Blood 2007;109:3936–44.
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308.
- López-Camarillo C, Lopez-Casamichana M, Weber C, Guillen N, Orozco E, Marchat LA. DNA repair mechanisms in eukaryotes: Special focus in Entamoeba histolytica and related protozoan parasites. Infect Genet Evol. 2009;9:1051–6.
- Le May N, Egly JM, Coin F. True lies: the double life of the nucleotide excision repair factors in transcription and DNA repair. J Nucleic Acids 2010;19:1–11.
- Liu H, Rudolf J, Johnson KA, McMahon SA, Oke M, Carter L, et al. Structure of the DNA repair helicase XPD. Cell 2008;133:801–12.
- Shuck SC, Short EA, Turchi JJ. Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology. Cell Res 2008;18:64–72.
- Osawa K. Gene polymorphisms and chemotherapy in non-small cell lung cancer. Chin J Lung Cancer 2009;12:837–40.
- Wang F, Chang D, Hu FL, Sui H, Han B, Li DD, et al. DNA repair gene XPD polymorphisms and cancer risk: a meta-analysis based on 56 case-control studies. Cancer Epidemiol Biomarkers Prev. 2008;17:507–17.
- Kuptsova N, Ambrosone CP, Weiss J, Baer MR, Sucheston LE, Zirpoli G, et al. XPD DNA nucleotide excision repair gene polymorphisms associated with DNA repair deficiency predict better treatment outcomes in secondary acute myeloid leukemia. Int J Mol Epidemiol Genet. 2010;1:278–94.
- Estey EH. Acute myeloid leukemia: 2013 update on risk-stratification and management. Am J Hematol. 2013;88(4):318–27. doi: 10.1002/ajh.23404.
- National Cancer Institute: Common terminology criteria for adverse events, v. 4.0. Available from: http://ctep.cancer.gov/reporting/ctc.html.
- Batar B, Güven M, Bariş S, Celkan T, Yildiz I. DNA repair gene XPD and XRCC1 polymorphisms and the risk of childhood acute lymphoblastic leukemia. Leuk Res. 2009;33(6):759–63. doi: 10.1016/j.leukres.2008.11.005 . Epub 2008 Dec 19.
- Heo SG, Hong EP, Park JW. Genetic risk prediction for normal-karyotype acute myeloid leukemia using whole-exome sequencing. Genom Inform. 2013;11(1):46–51. doi: 10.5808/GI.2013.11.1.46 . Epub 2013 Mar 31.
- Manuguerra M, Saletta F, Karagas MR, Berwick M, Veglia F, Vineis P, et al. XRCC3 and XPD/ERCC2 single nucleotide polymorphisms and the risk of cancer: a HuGE review. Am J Epidemiol. 2006;164(4):297–302 . Epub 2006 May 17.
- Crespan E, Garbelli A, Amoroso A, Maga G. Exploiting the nucleotide substrate specificity of repair DNA polymerases to develop novel anticancer agents. Molecules 2011;16(9):7994–8019. doi: 10.3390/molecules16097994.
- Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58(4):604–8.
- Spitz MR, Wu X, Wang Y, Wang LE, Shete S, Amos CI, et al. Modulation of nucleotide excision repair capacity by XPD polymorphisms in lung cancer patients. Cancer Res. 2001;61(4):1354–7.
- Rzeszowska-Wolny J, Polanska J, Pietrowska M, Palyvoda O, Jaworska J, Butkiewicz D, et al. Influence of polymorphisms in DNA repair genes XPD, XRCC1 and MGMT on DNA damage induced by gamma radiation and its repair in lymphocytes in vitro. Radiat Res. 2005;164(2):132–40.
- Shen M, Berndt SI, Rothman N, Demarini DM, Mumford JL, He X, et al. Polymorphisms in the DNA nucleotide excision repair genes and lung cancer risk in Xuan Wei, China. Int J Cancer 2005;116(5):768–73.
- Le Morvan V, Bellott R, Moisan F, Mathoulin-Pélissier S, Bonnet J, Robert J. Relationships between genetic polymorphisms and anticancer drug cytotoxicity vis-à-vis the NCI-60 panel. Pharmacogenomics 2006;7(6):843–52.
- Lunn RM, Helzlsouer KJ, Parshad R, Umbach DM, Harris EL, Sanford KK, et al. XPD polymorphisms: effects on DNA repair proficiency. Carcinogenesis 2000;21(4):551–5.
- Atkins CD. Single nucleotide polymorphisms and anthracycline cardiotoxicity in children: potential implications for adult oncology. J Clin Oncol. 2012;30(28):3563; author reply 3563–4. Epub 2012 Aug 27.
- Guven M, Guven GS, Oz E, Ozaydin A, Batar B, Ulutin T, et al. DNA repair gene XRCC1 and XPD polymorphisms and their association with coronary artery disease risks and micronucleus frequency. Heart Vessels 2007;22(6):355–60 . Epub 2007 Nov 26.
- Botto N, Masetti S, Petrozzi L, Vassalle C, Manfredi S, Biagini A, et al. Elevated levels of oxidative DNA damage in patients with coronary artery disease. Coron Artery Dis. 2002;13:269–274.
- Frosina G. Overexpression of enzymes that repair endogenous damage to DNA. Eur J Biochem. 2000;267:2135–2149.
- Qiu M, Yang X, Hu J, Ding X, Jiang F, Yin R, et al. Predictive value of XPD polymorphisms on platinum-based chemotherapy in non-small cell lung cancer: a systematic review and meta-analysis. PLoS One 2013;8(8):e72251. doi: 10.1371/journal.pone.0072251.
- Rumiato E, Cavallin F, Boldrin E, Cagol M, Alfieri R, Basso D, et al. ERCC1 C8092A (rs3212986) polymorphism as a predictive marker in esophageal cancer patients treated with cisplatin/5-FU-based neoadjuvant therapy. Pharmacogenet Genom. 2013;23(11):597–604.
