Abstract
Introduction
Acute myeloid leukemia (AML) is a highly heterogeneous disease, with biologically and prognostically different subtypes.
Aim
To study the impact of p53, p21, and mdm2 gene polymorphisms on the clinical outcome in adult AML patients treated at the National Cancer Institute (NCI) – Cairo University.
Methods
Forty-eight adult AML patients presented to the Medical Oncology Department, NCI, from April 2010 till November 2011. Clinical data and bone marrow samples were obtained. Molecular genetic analysis involving P53, MDM2, and P21 single-nucleotide gene polymorphisms was done using polymerase chain reaction-restriction fragment length polymorphism coupled analysis.
Results
The mean age was 35.7 years. After a median follow-up period of 12 months, 28 patients (58.4%) achieved complete remission (CR) and the overall survival (OS) was 8.7 months. Patients with homozygous Arg/arg at codon 72 of P53 had a better median OS months than Arg/Pro and Pro/Pro (13.4 vs. 8.4 vs. 1.5 months, respectively; P = 0.045). P53/p21 combination had a better median OS and disease-free survival (DFS) of 12.1 and 13.7 months for wild type cases (GG + Ser/ser) and 20.3 and 20.7 months for patients with either variant genes (GC + Ser/arg) compared with 1.1 and 1.9 months for patients with both variant genes (CC + arg/arg), (P = 0.037 and 0.004). The presence of wild genotype of either P21 or MDM2 may abolish the effect of P53 homozygous variant genotype on the OS. Neither p21nor mdm2 polymorphism alone showed an impact on OS or DFS. CR was not affected by any of the three gene polymorphisms.
Conclusion
The p53 pathway gene polymorphisms may affect the OS of adult AML patients.
Introduction
Acute myeloid leukemia (AML) is a highly heterogeneous disease, with biologically and prognostically different subtypes distinguished by cytogenetic and molecular genetics analyses. Application of high-throughout technologies including the whole genome sequencing of AML cases has revealed a plethora of recurrent mutational targets, including a number of genes encoding transcriptional regulators, not previously implicated in leukemogenesis. Deciphering the combinations of mutations that cooperate to induce AML and determining which particular alterations (or combinations) confer independent prognostic information represent the major ongoing challenges, which will necessitate the analysis of large cohort sizes involving international cooperation.Citation1 AML accounts for ∼25% of all leukemias in adults in the West and constitutes the most frequent form of leukemia. Worldwide, the incidence of AML is highest in the USA, Australia, and Western Europe.Citation2 The frequency of AML presentation in the National Cancer Institute (NCI), Cairo University during the years 2002–2003, was 1.9%, with a median age of 22 years and male-to-female ratio of 1.37. Leukemia ranks fourth among the most common sites among men and third among women.Citation3 The clinical outcome of AML is extremely variable, ranging from survival of a few days to cure. A number of clinical and biological features at presentation may predict the clinical outcome.Citation4 Cytogenetic abnormalities can be classified into three groups. The favorable group (10–15%), usually younger than 60 years, have translocations at t(8; 21) and t(16; 16) and an inversion on chromosome16. The unfavorable group, many of whom are elderly, have abnormalities that include monosomies and deletions in the long arms of chromosomes 5 and 7, as well as abnormalities that involve three or more chromosomes (i.e. a complex karyotype). The intermediate group (50–60%) have normal karyotypes and all other aberrations. Accordingly, two prognostic systems of classification have been proposed by the UK Medical Research Council (MRC)Citation5 and the US South West Oncology Group (SWOG)Citation6. The intermediate-risk category is the most heterogeneous group in AML. New molecular markers are being developed to further refine the prognostic groups, particularly patients with normal karyotypes. These markers include FLT3-ITD, nucleophosmin (NPM1), c-KIT, and CEBPA gene mutations.Citation7–Citation10 The dynamic processes of cell growth and division are under constant surveillance. As one of the primary ‘gatekeepers’ of the cell, the p53 tumor suppressor plays a major role in sensing and responding to a variety of stressors to maintain cellular homeostasis.Citation11 Under normal conditions, p53 levels are kept low, mainly through inhibition by MDM2.Citation12 There are three major outcomes of the p53 stress response: cell cycle arrest, cellular senescence, and apoptosis. Activated p53 binds DNA and activates the expression of several genes including WAF1/CIP1 encoding for p21. P21 functions as a regulator of cell cycle progression at S phase.Citation13 Single-nucleotide polymorphisms (SNPs) in codon Arg72Pro of P53 result in impairment of its tumor suppressor activity. A similar effect is caused by an SNP in codon 31 of P21. In contrast, an SNP in position 309 of MDM2 results in increased expression due to substitution of thymine by guanine. All three polymorphisms have been associated with increased risk of tumorigenesis.Citation14 Three days of an anthracycline (e.g. daunorubicin, idarubicin, or the anthracenedione mitoxantrone) and 7 days of cytarabine (100–200 mg/m2 continuous intravenous) currently remains the standard for induction therapy (‘3 + 7’ regimen). With such regimen, complete remission (CR) is achieved in 60–80% of younger adults.Citation15 Post-remission therapy with repetitive cycles of high-dose cytarabine (HiDAC; 3 g/m2 q12 hours on days 1, 3, and 5) is considered a reasonable choice for younger adult patients with CBF AML, and also for AML with mutated NPM1 without FLT3-ITD and with mutated CEBPA.Citation16 Allogeneic HSCT offers significant OS benefit for patients with intermediate- and high-risk AML.Citation17
Patients and methods
This retrospective study included 48 newly diagnosed adult primary AML patients (age 18–60 years) who presented to the Medical Oncology Department, NCI, Cairo University in the period between April 2010 and October 2011. The study protocol was approved by the Institutional Review Board (IRB) of the NCI. Cases were revised for thorough history and clinical examination, particularly for hepatomegaly, splenomegaly and lymphadenopathy, complete blood picture, and bone marrow aspiration (morphology, cytochemistry, immunophenotyping by flow cytometry, and cytogenetics). All of the patients were subjected to standard of care treatment at the National Cancer Institute. Induction therapy (‘3 + 7’) consisted of doxorubicin 45 mg/m2 for three consecutive days and ara-C 100 mg/m2 for 7 days. Patients who achieve CR were subjected to high-dose Ara-C plus mitoxantrone regimen (HAM; Ara-C 1500 mg/m2 q12 hours days 1–3, while mitoxantrone 12 mg/m2 days 3–5). All-trans-retinoic acid 45 mg/m2 and daunorubicin 60/m2 for 3 days were given to AML M3 cases.
Genotyping
MDM2 T309G genotyping
The MDM2 T309G polymorphism was determined using the method described by Hirata et al.Citation18 Each polymerase chain reaction (PCR) assay was performed using 100 ng of genomic DNA, 0.2 µM of each primer, 1 U of Hot Start Taq DNA polymerase (Quiagen, Hilden, Germany), 200 µM dNTP, 1.5 mM MgCl2, 10 mM Tris–HCl (pH 8.4), and 50 mM KCl. After initial denaturation for 10 minutes at 95°C, the PCR was performed for 35 cycles of 45 seconds at 95°C, 45 seconds at 59°C, and 1 minute at 72°C. The last elongation step was extended to 7 minutes. The amplified fragments targeted the site of polymorphism: the 158 bp fragment for MDM2 T309G containing the T → G base pair substitution at nucleotide 309 that creates an MspA1I restriction site. The digestion products were visualized with ethidium bromide after electrophoresis on 3.5% agarose gel at 100 V for 30 minutes. The MDM2 309TT wild-type homozygous was identified by the presence of only a 158 bp fragment. 309TG heterozygous was identified by 158, 112, and 46 bp fragments, and 309GG homozygous variant was identified by 112 and 46 bp fragments ().
Table 1. Primer sequence and PCR, PCR/RFLP fragment size
P53 codon arg72pro polymorphism
PCR was amplified in 25 µl containing 100 ng of genomic DNA, 0.5 µmol/l of primers (), 200 µmol/l dNTPs, 10 mmol/l Tris–HCl (pH 8.3), 2.5 mmol/l MgCl2, 50 mmol/l KCl, and 1 U of Hot Start Taq DNA polymerase (Quiagen). After initial denaturation for 10 minutes at 95°C, the PCR was performed for 35 cycles of 45 seconds at 95°C, 45 seconds at 58°C, and 1 minute at 72°C. The last elongation step was extended to 7 minutes. The Arg → Pro substitution abolishes a restriction site on digestion with BstUI restriction enzyme (10 U). The resulting restricted fragments were evaluated on a 3.5% agarose gel at 100 V for 30 minutes,Citation19 showing 113 and 86 bp bands for the wild type and 199, 113, and 86 bp bands for the heterozygous variant and the homozygous variant remains undigested 199 bp.
P21 codon 31 Ser/arg polymorphism
P21 codon 31 Ser/arg polymorphism was characterized by the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP).Citation18 DNA fragment of 225 bp was amplified in 25 µl containing 100 ng of genomic DNA, 0.5 µmol/l of primers, 200 µmol/l dNTPs, 10 mmol/l Tris–HCl (pH 8.3), 2.5 mmol/l MgCl2, 50 mmol/l KCl, and 1 U of Hot Start Taq DNA polymerase (Quiagen). After denaturation for 10 minutes at 95°C, the PCR was performed for 35 cycles of 1 minute at 95°C, 1 minute at 58°C, and 2 minute at 72°C. The last elongation step was extended to 7 minutes. The presence of polymorphic variant arg results in abolishing the restriction site odBlp I enzyme. So the PCR product (5–10 µl) was digested with Blp I (10 U, 37°C), and subjected to electrophoresis on a 2.5% agarose gel at 100 V for 30 minutes. The wild-type (ser/ser) results in two smaller fragments (122 and 103 bp) while the heterozygous variant result in 225, 122, 103 bp and the homozygous variant prevent digestion result in only 225 bp band.
Statistical methods
Data were analyzed using SPSS® statistical package version 20 for windows (SPSS Inc., Chicago, IL, USA). Numerical data were expressed as mean and standard deviation or median and range as appropriate. Qualitative data were expressed as frequency and percentage. Chi-square test (Fisher's exact test) was used to examine the relationship between qualitative variables. For quantitative data, comparison between two groups was done using either Student's t-test or Mann–Whitney test (non-parametric t-test) as appropriate. A P value <0.05 was considered significant. Kaplan–Meier method calculated all survival estimates. Other predictor and prognostic variables were related to survival using log-rank test. The P value was set at a significance level of 0.05.Citation20
Results
Patient characteristics
Out of 48 cases of AML, 22 were women (46%) and 26 were men (54%). The mean age was 35.7 years (SD = 11.49). More than half of the patients were AML-M1 and M2 subtypes (52%). Fever was encountered in 48% of the patients, hepatomegaly in 56%, and splenomegaly in 39%. Initial total leukocytic count (TLC) at diagnosis was <11 000/mm3 in 17%, between 11 000 and 50 000/mm3 in 50%, and >50 000/mm3 in 33%. Thirteen patients presented with a platelet count of <20 000 (27%). Bone marrow morphology was hypercellular in 90% of cases with a median blast percentage of 57% (ranging from 35 to 90%) ().
Table 2. Patient characteristics in the study group
Overall response and disease-free survival
At a median follow-up of 1 year, 28 of 48 patients (58.4%) achieved CR. Out of the 28 cases who achieved CR, 7 cases relapsed during the first year while 3 cases relapsed after 1 year and 3 cases died. The median overall survival (OS) was 8.7 months (ranging from 3.7 to 13.8). No correlation was found between hepatosplenomegaly, fever at initial presentation, initial leukocytic count, and CR or survival.
Impact of single gene polymorphism on clinical outcome in AML
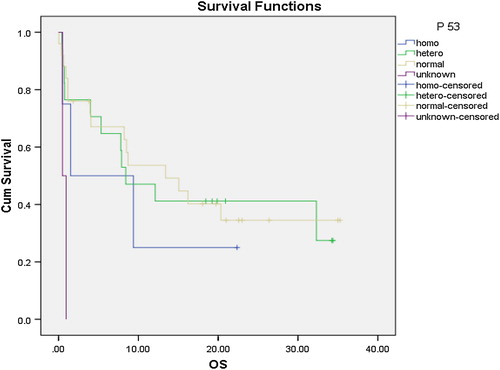
No significant correlation was found between single gene polymorphism and CR rates. Only P53 gene polymorphism had a significant effect on OS (), with the median OS for patients with P53 GG genotype, GC genotype, and CC genotype of 13.4, 8.4, 1.5 month, respectively (P = 0.045) (). No significant correlation was found between single gene polymorphism and disease-free survival (DFS) ().
Table 3. Impact of single gene polymorphism on CR
Table 4. Impact of single gene polymorphism on DFS and OS
Impact of combined gene polymorphism on clinical outcome in AML
Owing to the natural interaction of gene pathways in the human cell, combined effects of the three genes were compared with each of the outcome parameters in this study. Three combinations were measured (p53/p21, p53/mdm2, and mdm2/p21), and three outcome parameters were assessed (CR, DFS, and OS) in AML patients.
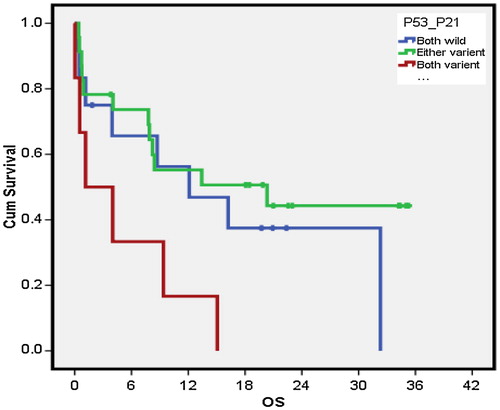
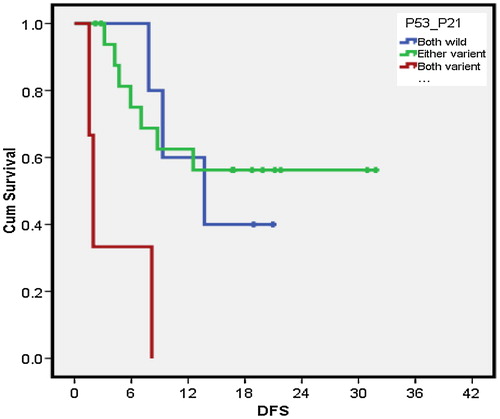
P53/p21 combination had a median OS of 12.1 months (0.3–23.8 months) for 12 cases that had both genes of wild type compared with 1.1 month (0–5.3 months) median OS for 6 patients with both genes of variant type. The either variant group showed a median OS of 20.3 months (0–43.8 months) for a total of 23 cases (P = 0.037). Owing to the significant value, pairwise comparison of p53/p21 combination was done. The either variant patients had a better median OS than both the variants (P = 0.015) (). On the other hand, neither p53/mdm2 combinations nor mdm2/p21combinations had significant impact on OS (). P53/p21 combination showed a median DFS of 13.7 (4.2–23.1 months) months for 12 cases that had both genes of wild type compared with 1.9 months (1.2–2.6 months) for 6 patients with both genes of variant type (P = 0.004). The either variant group showed a mean DFS of 20.7 (SD = 14.9) months for a total of 23 cases. Owing to the significant value, pairwise comparison of p53/p21 combination was done. The either variant group had a better DFS compared with both variant groups (P = 0.003). Moreover both wild-type groups had a better DFS compared with both variant groups (P = 0.022) (). On the other hand, neither p53/mdm2 combinations nor mdm2/p21combinations had significant impact on DFS ().
Table 5. Impact of combined gene polymorphisms on OS and DFS
None of the three different gene polymorphism combinations p53/p21, p53/mdm2, and mdm2/p21 had a significant impact on CR rates ().
Table 6. Impact of combined gene polymorphisms on CR
Discussion
AML accounts for ∼25% of all leukemias in adults in the West and constitutes the most frequent form of leukemia. Worldwide, the incidence of AML is the highest in the US, Australia, and Western Europe.Citation2 At the National Cancer Institute (NCI) Cairo University during the years 2002–2003, out of a total of 18 496 new cancer cases, 349 patients (1.9%) were diagnosed as AML with a median age of 22 years and a male-to-female ratio of 1.37. Leukemia ranked the fourth most common site among men and third among women.Citation3In this study, a slight male predominance was observed with a male-to-female ratio of 1.18/1, similar to that reported by El-Zawahry et al.Citation21 (1.05/1). Age ranged, in our study, between 20 and 60 years with a mean of 35.7 years. Similar age at diagnosis was reported by El-Zawahry et al.Citation21 (33.87 ± 12.3 years), but Zolota et al.Citation22 reported a higher age range (from 42 to 86 years with a median of 70 years).The overall CR rate was 58.3% with a relapse rate of 35.7%. This is to be compared with the retrospective study by Eastern Cooperative Oncology Group (ECOG), where a CR rate of 62% was attained, but 76% of these patients have relapsed or died, the OS rate at 5 years was 15%.Citation23 Out of the 10 cases of M3, 6 cases showed SNP for MDM2 T309G genotyping (T-G or G-G), while only 2 cases showed SNP for P21 codon 31 Ser/arg polymorphism (Ser-Arg or Arg-Arg), and 3 cases were polymorphic for p53 arg72pro gene (G-C or C-C). Noticeably, 7 out of the 10 cases of M3 achieved CR (70%) and were alive till the end of the median observation period (12 months) in spite of the variable SNPs discovered for the three genes. The other three cases who failed to achieve CR had no SNP for mdm2 or p53 while two of them had P21 codon 31 Ser/arg polymorphism. Our CR rate for M3 is comparable to 80% CR rate reported by Fenaux et al.Citation24 In our study, P53 gene polymorphism was done on 46 of 48 cases of AML, P53 GG, P53 GC, and P53 CC genotypes were identified in 54.3, 36.9, and 8.6% respectively, compared with 45.4, 31.2, and 23.4% respectively, reported by Dunna et al.,Citation25 and with 22.5, 55, and 22.5%, respectively, reported by Xiong et al.Citation26 Mdm2 gene polymorphism was performed on 45 of 48 cases of AML, MDM2 309TT, MDM2 309TG, and MDM2 309GG were identified in 28.9, 48.9, and 22.2%, respectively, compared with 13.9, 53.2, and 32.9%, respectively, reported by Xiong et al.Citation26 P21 gene polymorphism revealed 62.8% for P21 serine (p21-Ser) homozygous, 4.6% for P21 arginine (p21-Arg) homozygous, and 32.5% for P21 serine/arginine (p21-Ser/Arg) heterozygous. We investigated for an association of polymorphism of the three genes with clinical outcome. Polymorphism of each gene alone showed that p53 significantly affected OS while the other two genes failed to show a significant impact on survival. None of the three single gene polymorphisms or different gene polymorphism combinations: p53/p21, p53/mdm2, and mdm2/p21 had a significant impact on CR rates. The median OS was 13.4, 8.4, 1.5 months for homozygous arginine p53 genotype (GG), heterozygous variant of p53 gene (GC), and homozygous proline p53 genotype (CC), respectively (P = 0.045). In a similar study by Nakano et al.,Citation27 they concluded that p53polymorphism at codon 72 was not associated with risk, pathophysiology, or therapeutic response of AML but reported that p53 mutation was an independent factor for short OS. Moreover Shi et al.,Citation28 reported that post-chemotherapy response analysis in AML patients GG homozygous for TP53 C > G were independently linked to inferior treatment outcomes. The arginine (Arg 72) allele increases the ability of p53 to locate to the mitochondria and induce cellular death, whereas proline allele (Pro 72) exhibits a lower apoptotic potential and an increased cellular arrest in G1 of the cell cycle.Citation29 In colony-survival assays performed in H1299 cells, the cytotoxicities of cisplatin and doxorubicin were higher in 72R-expressing cells than in 72P-expressing cells. Similarly, there was reproducibly higher cytotoxocity of etoposide, 5-FU, and taxol in cells expressing 72R p53.Citation30 Other forms of P53 change than polymorphism reported significantly shorter OS of patients with p53 gene mutations than patients without mutations,Citation31 also p53 deletion without multiple aberrations was an independent negative prognostic factor for DFS (P < 0.001), relapse risk (P = 0.028), and OS (P < 0.001) as reported by Seifert et al.,Citation32 and negative prognostic impact of p53 mutations, or p53 mutations, and 17p loss of heterozygosity combined on survival in AML reported by Parkin et al.Citation33 Moreover a similar result of significantly different OS and PFS between cases retaining a wild-type 72R allele, cases retaining a wild-type 72P allele, and cases retaining both wild-type 72R and 72P alleles with the best prognosis in cases retaining a wild-type 72R allele in advanced head and neck squamous cell carcinomas.Citation30 The median OS was 9.3, 15, and 0.9 months for P21 serine (p21-Ser) homozygous, P21 serine/arginine (p21-Ser/Arg), and P21 arginine (p21-Arg) homozygous, respectively (P = 0.310), the same conclusion of the absence of OS significance for P21 protein expression was reported by others.Citation22 Previous studies have detected very low levels of p21 protein in freshly isolated AML cells, but a strong and rapid induction of p21 when AML cells were exposed to daunorubicin.Citation34 The p53 analysis when combined with p21 showed a striking significance on overall and DFS of AML (P = 0.037 and 0.004, respectively). These results support the fact that p21 plays an important role on tumor suppression but gene variation fails to translate this effect on clinical basis, however, when p21 polymorphism is present with a p53 variant gene it has a synergistic effect clinically with a negative impact on DFS (P = 0.004) and OS (P = 0.037). MDM2 has been found to be frequently overexpressed in AML, and can enhance the tumorigenic potential and resistance to apoptosis through abrogation of p53 function.Citation35 In our study, mdm2 polymorphism failed to show clinical significance, the median OS was 13.4, 7.8, 16.2 months for patients with MDM2 309TT, MDM2 309TG, and MDM2 309GG, respectively (P = 0.846), the same conclusion was reported by othersCitation22,Citation36 for MDM2 protein expression. It was reported that, compared with the T allele, the variant G allele increases the affinity of the transcription factor SP1 to the MDM2 promoter and enhances the expression of MDM2 RNA and MDM2 protein, thereby leading to attenuation of the p53 stress response.Citation37 Mdm2 polymorphism was extensively analyzed in terms of clinical significance and the risk of cancer in many solid malignancies. In almost all of the studies, mdm2 polymorphism was associated with poor prognosis. MDM2 overexpression was an independent predictor for poor prognosis of advanced gastric cancer,Citation38 and colorectal cancer.Citation39 The interactive effect between p53 and mdm2 gene polymorphisms that was detected such that MDM2 TT TP53Arg/Arg double homozygotes, and individuals carrying both an MDM2 G allele and a TP53 Pro allele, were at increased risk of t-AML in patients previously treated with chemotherapyCitation40 was not reproduced on clinical outcome in this study which may be due to the small sample size. Combined analysis showed that patients carrying the variant P53 Pro/Pro-MDM2 GG genotypes had a survival time of only half of that for those carrying the wild-type genotypes (P = 0.039) in non-small cell lung cancer (NSCLC) patients.Citation41 Moreover MDM2 T/G + G/G genotypes, and more than two of the total variant alleles in TP53 and MDM2, were independently associated with improved cancer-specific survival (P = 0.031 and P = 0.015, respectively) in patients with bladder cancer receiving platinum-based chemoradiotherapy. The combined effect of TP53 R72P and MDM2 SNP309 on survival was worse.Citation42 No impact of combined mdm2 and p21 polymorphisms on clinical outcome in AML was observed. In our study, no significant relationship was found between mdm2/p21 polymorphism and OS (P = 0.158) or DFS (P = 0.112).
This supports the fact that P53 is an independent prognostic factor of survival. None of the three genes, separately, had an impact on DFS or CR rates.
Disclaimer statements
Contributors MMS collected cases data; GTE performed gene polymorphism studies; MEG treated cases and wrote the paper; OMA was responsible for the statistical analysis; TMAH revised and finalized the data.
Funding None.
Conflicts of interest None.
Ethics approval The study was approved by the Ethics Committee of the Egyptian NCI, Cairo University.
References
- Grimwade D, Hills RK. Independent prognostic factors for AML outcome. Hematol Am Soc Hematol Edu Program 2009;1:385–95.
- Döhner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, et al. Diagnosis, management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European Leukemia-Net. Blood 2012;115:453–74.
- Elattar I. (2005): Cancer in the Arab World: Magnitude of the Problem. UICC March, 2005 [Online]. [accessed 2011 Jul 5] Available from: http://www.nci.edu.eg/lectures/cancer_problem/Cancer%20in%20the%20Arab%20World%20UICC%20Mar24-05.pdf
- Avivi I, Rowe JM. Prognostic factors in acute myeloid leukemia. Curr Opin Hematol. 2005;12:62–7.
- Grimwade D, Walker H, Harrison G, et al. The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood 2001;1(98):1312–20.
- Slovak ML, Kopecky KJ, Cassileth PA, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood 2000;96(13):4075–83.
- Ferrara F, Palmieri S, Pollio F, et al. Presence of FLT3 mutations does not impair stem cell mobilization and feasibility of autologous peripheral blood stem cell transplantation in acute myeloid leukemia. Biol Blood Marrow Transplant. 2006;12:981–6.
- Dohner K, Schlenk RK, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood 2005;106:3740–6.
- Cairoli R, Beghini A, Grillo G, et al. Prognostic impact of c-KIT mutations in core binding factor leukemias: an Italian retrospective study. Blood 2006;107:3463–8.
- Marcucci G, Maharry K, Radmacher MD, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B Study. J Clin Oncol. 2008;26:5078–87.
- Miliani de Marval PL, Zhang Y. The RP-Mdm2-p53 pathway and tumorigenesis. Oncotarget 2011;2(3):234–8.
- Pan W, Issaq S, Zhang Y. The in vivo role of the RP-Mdm2-p53 pathway in signaling oncogenic stress induced by pRb inactivation and Ras overexpression. PLoS ONE 2011;6(6):e21625.
- Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65(10):3980–5.
- Onel K, Cordon-Cardo C. MDM2 and prognosis. Mol Cancer Res. 2004;2:1–8.
- Estey E, Döhner H. Acute myeloid leukaemia. Lancet 2006;368(9550):1894–907.
- Schlenk RF, Dohner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–18.
- Koreth J, Schlenk R, Kopecky KJ, et al. Allogeneic stem cell transplantation for acute myeloid leukemia in first complete remission: systematic review and meta-analysis of prospective clinical trials. JAMA 2009;301(22):2349–61.
- Hirata H, Hinoda Y, Kikuno N, et al. MDM2 SNP 309 polymorphism as risk factor for susceptibility and poor prognosis in renal cell carcinoma. Clin Cancer Res. 2007;13:4123–9.
- Ara S, Lee PS, Hansen MF, Saya H. Codon 72 polymorphism of the TP53 gene. Nucleic Acids Res. 1990;18:496.
- Kleinbaum DG, Klein M. Survival analysis: a self learning text. 2nd ed. New York, NY: Springer; 2005.
- El-Zawahry HM, Zeeneldin AA, Samra MA, Mattar MM, El-Gammal MM, Abd El-Samee A, et al. Cost and outcome of treatment of adults with acute myeloid leukemia at the National Cancer Institute-Egypt. J Egypt Natl Canc Inst. 2007;19(2):106–13.
- Zolota V, Sirinian C, Melachrinou M, Symeonidis A, Bonikos DS. Expression of the regulatory cell cycle proteins p21, p27, p14, p16, p53, mdm2, and cyclin E in bone marrow biopsies with acute myeloid leukemia. Correlation with patients’ survival. Pathol Res Pract. 2007;203:199–207.
- Bennett JM, Young ML, Andersen JW, et al. Long-term survival in acute myeloid leukemia: the Eastern Cooperative Oncology Group experience. Cancer 1997;80 (11 Suppl):2205–9.
- Fenaux P, Mufti GJ, Hellstrom-Lindberg E, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10(3):223–32.
- Dunna NR, Vure S, Sailaja K, Surekha D, Raghunadharao D, Rajappa S, et al. TP53 codon 72 polymorphism and risk of acute leukemia. Asian Pac J Cancer Prev. 2012;13:347–50.
- Xiong X, Wang M, Wang L, Liu J, Zhao X, Tian Z, et al. Risk of MDM2 SNP309 alone or in combination with the p53 codon 72 polymorphism in acute myeloid leukemia. Leuk Res. 2009;33:1454–8.
- Nakano Y, Kiyoi H, Naoe T, et al. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood 1999;93:3074.
- Shi JY, Ren ZH, Jiao B, Xiao R, Yun HY, Chen B, et al. Genetic variations of DNA repair genes and their prognostic significance in patients with acute myeloid leukemia. Int J Cancer 2011;128(1):233–8. doi: 10.1002/ijc.25318.
- Bergamaschi D, Samuels Y, Sullivan A, et al. iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat Genet. 2006;38:1133–41.
- Sullivan A, Syed N, Gasco M, Bergamaschi D, Trigiante G, Attard M, et al. Polymorphism in wild-type p53 modulates response to chemotherapy in vitro and in vivo. Oncogene 2004;23:3328–37.
- Melo MB, Ahmad NN, Lima CS, Pagnano KB, Bordin S, Lorand-Metze I, et al. Mutations in the p53 gene in acute myeloid leukemia patients correlate with poor prognosis. Hematology 2002;7(1):13–9.
- Seifert H, Mohr B, Thiede C, Oelschlägel U, Schäkel U, Illmer T, et al. The prognostic impact of 17p (p53) deletion in 2272 adults with acute myeloid leukemiap53 deletion: a high-risk factor in AML. Leukemia 2009;23:656–63. doi:10.1038/leu.2008.375.
- Parkin B, Erba H, Ouillette P, Roulston D, Purkayastha A. Acquired genomic copy number aberrations and survival in adult acute myelogenous leukemia. Blood 2010;116:4958–67.
- Radosevic N, Delmer A, Tang R, Marie J-P, Ajchenbaum-Cymbalista F. Cell cycle regulatory protein expression in fresh acute myeloid leukemia cells and after drug exposure. Leukemia 2001;15:559–66.
- Moll UM, Petrenko O. The MDM2-p53 interaction. Mol Cancer Res. 2003;1:1001–8.
- Faderl S, Kantarjian HM, Estey E, Manshouri T, Chan CY, RahmanElsaied A, et al. The prognostic significance of p16(INK4a)/p14(ARF) locus deletion and MDM-2 protein expression in adult acute myelogenous leukemia. Cancer 2000;89(9):1976–82.
- Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53tumor suppressor pathway and accelerates tumor formation in humans. Cell 2004;119(5):591–602.
- Ohmiya N, Taguchi A, Mabuchi N, et al. MDM2 promoter polymorphism is associated with both an increased susceptibility to gastric carcinoma and poor prognosis. J Clin Oncol. 2006;24:4434–40.
- Yu Y, Chen GH, Hu J, et al. The expression and the biological significance of mdm2 in human colorectal adenocarcinoma. Chin J Clin Oncol (Chinese) 2006;33:264–6.
- Ellis NA, Huo D, Yildiz O, Worrillow LJ. MDM2 SNP309 and TP53 Arg72Pro interact to alter therapy-related acute myeloid leukemia susceptibility. Blood 2008;112:741–9.
- Liu L, Wu C, Wang Y, Zhong R, Duan S, Wei S, et al. Combined effect of genetic polymorphisms in P53, P73, and MDM2 on non-small cell lung cancer survival. J Thorac Oncol. 2011;6(11):1793–800. doi: 10.1097/JTO. 0b013e3182272273.
- Schmidt MK, Tommiska J, Broeks A, van Leeuwen FE, Van't Veer LJ, Pharoah PDP, et al. Combined effects of single nucleotide polymorphisms TP53 R72P and MDM2 SNP309, and p53 expression on survival of breast cancer patients. Breast Cancer Res. 2009;11:R89. doi:10.1186/bcr2460.