Abstract
Background
Calreticulin (CALR) mutations were recently identified in a substantial proportion of persons with essential thrombocythemia (ET) and with primary myelofibrosis (PMF) without JAK2V617F. Consequently rapid, sensitive, and specific methods to detect and quantify these mutations are needed.
Methods
We studied samples from 1088 persons with myeloproliferative neoplasms (MPNs) including 421 JAK2V617F negative subjects with ET, PMF, polycythemia vera (PV), chronic myeloid leukemia (CML) and hyper-eosinophilic syndrome (HES). Detection of CALR exon 9 mutations was done by PCR amplification followed by fragment length analysis and direct sequencing. Dilution assays were used to determine CALR mutant allele burden.
Results
We detected CALR mutations in blood and bone marrow samples from 152 subjects with ET and with PMF but not in samples from normal or persons with PV, CML, or HES. CALR mutant peaks were distinct from wild-type peaks and dilution experiments indicated a sensitivity level of 0.5–5% for a CALR mutant allele in a wild-type background. Diverse types of mutations were detected including deletions, insertions, and complex indels. All mutations were confirmed by direct sequencing. We also used dilution experiments to quantify mutant allele burden. We were able to reproducibly detect mutant allele levels as low 5% (0.5–5%) in a wild-type background.
Conclusions
PCR amplification followed by fragment length analysis is a rapid, sensitive, and specific method for screening persons with MPNs for CALR mutations, especially those with ET and PMF and for estimating mutant allele burden.
Introduction
JAK2V617F mutations are present in 50–60% of persons with essential thrombocythemia (ET) and with primary myelofibrosis (PMF).Citation1 Mutations in MPL codon 515 are present in another 3–5%.Citation2–Citation8 Recently, mutations in CALR were reported in 30–70% of persons with ET and PMF without mutations in JAK2V617F or MPL.Citation9,Citation10 CALR mutations are confined to exon 9 with loss of most of the C-terminal acidic domain, multiple calcium binding sites, and the KDEL sequence and are thought to be early events in MPN development.Citation10 Almost all mutations are insertions and deletions resulting in a shift of one base pair in the reading frame. Most of the mutants correspond to a 52 bp deletion (L367fs*46) and a 5 bp TTGTC insertion (K385fs*47).Citation9–Citation14 These frame-shift mutations produce a significantly altered C-terminal domain.Citation10 The role of CALR mutations in the etiology of ET and PMF is unknown and correlates between CALR mutation mutant allele burden and clinical features are being studied. It is likely CALR mutation state will soon be added to the WHO diagnostic criteria for ET and PMF.Citation12,Citation13
The detection of CALR mutations in most of the published reports is done by PCR followed by Sanger sequencing.Citation9,Citation10 However, direct sequencing is expensive, time-consuming, and sensitivity is only about 10% for a mutated allele in a wild-type background. Consequently, there is a need for a rapid, sensitive, and specific test to detect CALR mutations. PCR followed by fragment length analysis is widely used in other settings because it is simple, rapid and low cost. We studied feasibility of using PCR followed by fragment length analysis for the detection of CALR mutation in subjects with ET, PMF and PV without mutations in JAK2V617F.
Methods
Subjects and samples
We studied 1088 persons with myeloproliferative neoplasms (MPNs) diagnosed in 2006–2013 at People's Hospital, Beijing, China, including 614 with ET, 132 with PMF, 283 with polycythemia vera (PV), 49 with chronic myeloid leukemia (CML), and 10 with hyper-eosinophilic syndrome (HES). We also studied 20 normal persons. Diagnoses were based on the revised World Health Organization criteria.Citation15 JAK2V617F mutation was analyzed using the RQ-PCR with TaqMan MGB probes.Citation16 Subjects with ET or PMF without JAK2V617F were studied for CALR exon 9 mutations. Six hundred and sixty-seven of the 1088 subjects had JAK2V617F. Frequencies of JAK2V617F were 88% in PV, 57% in ET and 53% in PMF. The 421 subjects without JAK2V617F composed of 265 ET, 62 PMF, 35 PV, 49 CML, and 10 HES. Written informed consent was obtained from all subjects and study-design adhered to the principles of the Helsinki Declaration and was approved by the Ethics Committee of People's Hospital.
Mononuclear cells were isolated from bone marrow (n = 846) and blood samples (n = 242) by Ficoll-Hypaque density-gradient centrifugation. DNA was extracted using the DNAzol kit (Invitrogen, Carlsbad, CA, USA) and quantified using the NanoDrop technology (ND-1000 spectrophotometer; NanoDrop Technology, Wilmington, DE, USA). All samples used for PCR had 260 nm:280 nm absorbance ratios >1.8.
PCR fragment analyses
Exon 9 region of human CALR gene was amplified for fragment analyses. PCR was performed using DNA from the samples as the template and primers as described.Citation10 The forward primer was 5′-FAM-CCTGCAGGCAGCAGAGAAAC-3′ and reverse primer was 5′-ACAGAGACATTATTTGGCGCG-3′. Another pair of primers chosen by Klampfl et al.Citation9 was used as a control; 25 μl of the PCR mixture contained 12.5 μl of 2 × Universal PCR Master mix (TianGen Biotech Co., Beijing, China), 400 nM primers and 300 ng DNA. The PCR protocol involved denaturing at 95°C for 5 minutes followed by 36 cycles at 95°C for 40 seconds, 55°C for 40 seconds, and 72°C for 1 min and a final extension at 72°C for 10 min. PCR products were mixed with a size marker (GeneScan™-600 LIZ@ Size Standard v2.0; Applied Biosystems, Foster City, CA, USA) and incubated for 5 minutes at 95°C. Mixtures were then analyzed by capillary electrophoresis on an ABI 3500 Genetic Analyzer using 36-cm capillaries and POP-7 polymer. Data were analyzed using the GeneMapper v4.1 software (Applied Biosystems, Foster City, CA, USA). Sensitivity of PCR fragment analysis was determined by testing a mixture of plasmids containing either CALR exon 9 wild-type or type-1 mutation (L367fs*46) in varying proportions.
CALR gene sequencing
Exon 9 region of human CALR gene was amplified for CALR mutational analysis. Fifty microlitres of the PCR mixture contained 25 μl of 2 × Universal PCR Master mix (TianGen Biotech Co., Beijing, China), 400 nM primers and 300 ng DNA. PCR products were generated as described above. Purified PCR products were analyzed with an ABI3700 DNA sequencer.
Preparation of CALR plasmid standards
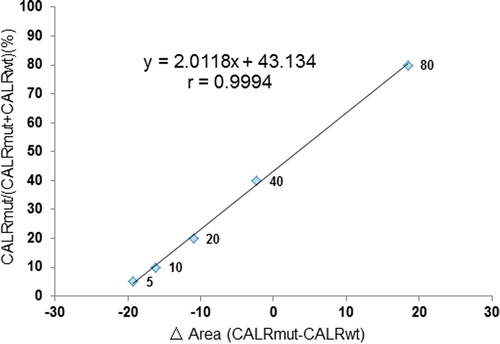
The 285 bp fragment of CALR (wild-type sequence) and the 233 bp fragment (type-1 mutation, p.L367fs*46) of CALR from subjects with the CALR mutant were sub-cloned into the pMD18-T vector (Takara, Dalian, China). Transformation, screening, and sequencing were done to obtain CALR wild-type sequence and CALR mutation sequence plasmid standards. Copy number was calculated based on the optical density value and standards serially diluted 10-fold (10E + 2–10E + 7 copies) to plot a standard curve. For diagnoses, we performed progressive dilutions of mutant plasmid alleles from 0.1 to 80% in a solution of wild-type plasmid alleles as in the non-homogeneous DNA samples. Mean area values (area CALRmut– area CALRwt) were calculated and percent mutant alleles in the sample computed by comparison with a standard curve of serial dilutions of mutant plasmid mixtures in wild-type plasmid DNA.
Results
Standard curve for detecting CALR mutant allele burden and sensitivity
Plasmid standards of CALR wild-type and CALR mutated-type 1 were prepared. Plasmid DNA concentration was determined by reading the absorbance. Six serial 10-fold plasmid dilutions (10E + 2–10E + 7 copies) were amplified by PCR in constructing a standard curve for the assessment of copy number. The CALR wild-type and CALR mutated plasmids had correlation coefficients >0.99. In subjects, we also analyzed progressive dilutions of each mutant plasmid allele from 0.1 to 80%, in a solution of wild-type plasmid allele as in the non-homogeneous DNA samples. Mean area values (area CALRmut−area CALRwt) were calculated and percent mutant alleles calculated by comparison to a standard curve of serial dilutions of mutant plasmid mixtures in wild-type plasmid DNA. Standard curves for detecting mutant allele burden were: y = 2.0118x + 43.134; r = 0.9994. Correlation coefficients were >0.99 (). Detection sensitivity was 0.5–5% for a mutated allele in a wild-type background. Coefficient of variation of the area value at each dilution was <5%. To determine whether similar sensitivity could be obtained in clinical samples, dilution experiments were also performed using six DNA samples of mutated-type-1 or -2 subjects in a wild-type control DNA sample. In all subjects, the mean area value of triplicate determinations (area CALRmut−area CALRwt) was in the linear range suggesting >5% (0.5–5%) mutated allele burden was reproducibly detected in clinical DNA samples. High correlation coefficients (>0.99) allowed an accurate assessment of the quantity of mutation in test samples.
Mean area values (area CALRmut−area CALRwt) were calculated and percent mutant alleles estimated by comparison to a standard curve of serial dilutions of mutant plasmid mixtures in wild-type plasmid DNA. Reference curve equation for detection of mutant allele burden
Sensitivity of CALR mutation screening by sequencing and fragment analysis
Different proportions of CALR exon 9 plasmids wild-type and mutant (L367fs*46) were mixed and genotyped. CALR mutation proportions were 0.5, 2.5, 5, 10, 20, 40, 60 and 100%. Detection sensitivity of direct sequencing was >10% for mutated CALR allele in a wild-type background. Fragment analysis traces showed the mixed wild-type and mutant peaks were clear down to a 5% mutant allele fraction (as shown in ). High correlation coefficients (>0.99) permitted accurate assessment of the quantity of CALR mutations in unknown samples.
Figure 2. Titration analyses of sensitivity of CALR mutation screening by sequencing and fragment analyses.
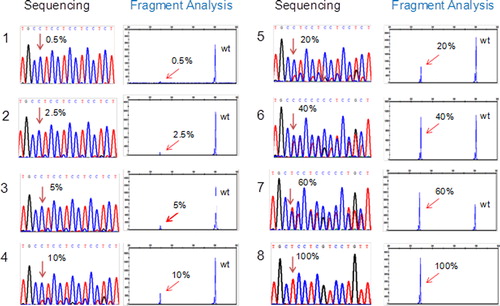
CALR exon 9 plasmids wild-type and mutant (L367fs*46) were mixed and genotyped. Detection sensitivity of direct sequencing was >10% for mutated CALR allele in a wild-type background. Traces of fragment analysis show the wild-type and mutant is distinguishable at the 5% mutant allele fraction.
Detection of CALR mutation by fragment analysis
One hundred and fifty-two samples showed CALR mutant peaks from wild-type peaks, including 124 (20%; 95% confidence interval (CI), 17–24%) patients with 614 ET and 28 (21%, (16–29%)) with 132 PMF. No CALR mutations were detected in the 35 JAK2V617F negative subjects with PV, 49 subjects with CML, 10 subjects with HES and 20 normals.
CALR mutation sequencing
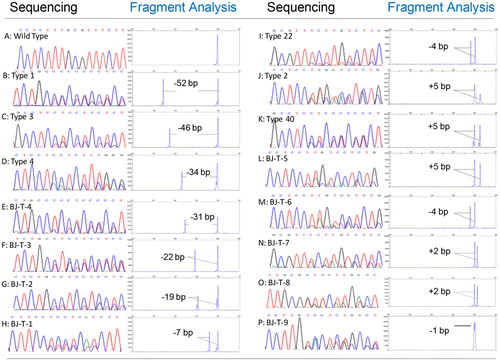
Sequence analysis confirmed all CALR mutations were heterozygous for an insertion, a deletion or complex indels in exon 9. One hundred and fifty-two CALR mutations were identified including eight deletions, three insertions and four complex indels (as shown in ). A 52-bp deletion (type-1 mutation) and a 5-bp insertion (type-2 mutation) were the most frequent alterations detected in 77 (51%, (42–59%)) and 53 (35%, (28–44%)) of subjects with mutations.
Figure 3. Sequencing traces show heterozygous mutation of CALR. Gene scan electropherogram from the PCR method and partial sequence of CALR exon 9 from the sequencing method (numbering according to GenBank access number: NC_000019.9). A-P: Detected a wild-type and 15 CALR mutation types by sequencing and fragment analysis methods. A: wild-type, B-I: deletions, H-L: insertions, M-P: complex indels.
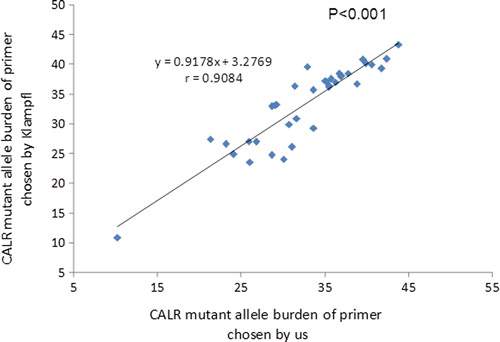
Comparison of primers
We compared data using our primers with those of Klampfl et al. using different primers for 15 CALR types, detection rate, and percent mutant alleles. Results were similar with detection rates of 52% (95% CI, 40–64%) vs. 52% (95% CI, 40–64%) P > 0.05). The correlation coefficient of percent mutant alleles is 0.9084 (. P < 0.001).
Discussion
CALR mutations were recently identified in a substantial proportion of persons with ET and PMF without JAK2V617F or MPL mutations.Citation9,Citation10 Most studies analyzed CALR mutations by PCR followed by Sanger sequencing.Citation9–Citation11,Citation14,Citation17,Citation18 A recent report proposed analysis by PCR followed by fragment analysis.Citation19 We developed a rapid highly sensitive and specific fragment method which allows the identification of exon 9 CALR mutations. No false-positives or -negatives were observed and the fragment method with a detection sensitivity of 0.5–5% of mutated DNA in wild-type background. This is about 2-fold as sensitive as Sanger sequencing. Correlation coefficients >0.99 allowed precise quantification of CALR mutations in unknown samples.
Using our technique we detected 15 types of mutations in 152 CALR (insertions and deletions) mutations including eight deletions, three insertions and four complex indels. Most previously described CALR mutations are insertions/deletions. A 52 bp deletion (L367fs*46) and a TTGTC insertion (K385fs*47) constitute about 85% of reported CALR mutations.Citation9,Citation10,Citation14,Citation17 In our study, the peak pattern of the 52 bp deletion was distinct from the 5 bp insertion. Two basepair insertions other than TTGTC are also described and sequencing would be needed to between these if precise knowledge of the details of the insertion were clinically important. One basepair change is unlikely to be distinguished only by fragment analysis. In our study, peak pattern of the 1 bp deletion was ambiguous and a mean area value could not be calculated. However, two peak areas of each sample were approximated such that we could estimate mutant allele burden at about 40%. Although our data are similar Klampfl et al. with regard to detecting a CALR mutation as a binary, they differ in that we were able to estimate mutant allele burden. Correlation coefficient of percent mutant alleles is about >0.9.
Others recently reported an assay to detect CALR mutations using high-resolution melting analysis followed by real-time PCR.Citation20 However, this assay can quantify only the two most common mutations in CALR, whereas our assay detects all CALR mutations except deletion or insertion of 1 bp. Consequently, our assay is more widely applicable to testing of persons with ET and PMF.
CALR mutation is suggested as a criteria in the upcoming revision of 2008 WHO diagnostic criteria for ET and PMF.Citation12,Citation13 PCR followed by fragment length analysis is a rapid, highly sensitive and specific low-cost technique to screen for CALR mutations and estimate mutant allele burden. These features make it potentially useful for diagnosis and monitoring therapy-response.
Acknowledgment
RPG acknowledges support from the NIHR Biomedical Research Centre funding scheme.
Disclaimer statements
Contributors GRR, SSC and RPG designed the project and prepared the typescript. QMY, ZJ and NL provided data, performed statistical analyses and contributed to the typescript. JLL and LDL performed technical details and provided clinical data. All authors approved the typescript.
Funding Supported by grants from the National Basic Research Program of China (grant 2013CB733701), the Key Program of National Natural Science Foundation of China (grant 81230013), the National Natural Science Foundation of China (grant 81170484), the Beijing Municipal Natural Science Foundation (grant 7122199) and the Specialized Research Fund for the Doctoral Program of Higher Education of China (grant 20130001110079).
Conflicts of interest None.
Ethics approval Written informed consent was obtained from all subjects and study-design adhered to the principles of the Helsinki Declaration and was approved by the Ethics Committee of People's Hospital.
References
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: IARC; 2008.
- Campbell PJ, Scott LM, Buck G, Wheatley K, East CL, Marsden JT, et al. Definition of subtypes of essential thrombocythaemia and relation to polycythaemia vera based on JAK2 V617Fmutation status: a prospective study. Lancet 2005;366:1945–53.
- Antonioli E, Guglielmelli P, Pancrazzi A, Bogani C, Verrucci M, Ponziani V, et al. Clinical implications of the JAK2 V617F mutation in essential thrombocythemia. Leukemia 2005;19:1847–9.
- Tefferi A, Lasho TL, Schwager SM, Steensma DP, Mesa RA, Li CY, et al. The JAK2 V617F tyrosine kinase mutation in myelofibrosis with myeloid metaplasia: lineage specificity and clinical correlates. Br J Haematol. 2005;131:320–8.
- Barosi G, Bergamaschi G, Marchetti M, Vannucchi AM, Guglielmelli P, Antonioli E, et al. JAK2 V617f mutational status predicts progression to large splenomegaly and leukemic transformation in primary myelofibrosis. Blood 2007;110:4030–6.
- Tefferi A, Lasho TL, Huang J, Finke C, Mesa RA, Li CY, et al. Low JAK2V617F allele burden in primary myelofibrosis, compared to either a higher allele burden or unmutated status, is associated with inferior overall and leukemia-free survival. Leukemia 2008;22:756–61.
- Pardanani AD, Levine RL, Lasho T, Pikman Y, Mesa RA, Wadleigh M, et al. MPL515 Mutations in myeloproliferative and other myeloid disorders: a study of 1182 patients. Blood 2006;108:3472–6.
- Guglielmelli P, Pancrazzi A, Bergamaschi G, Rosti V, Villani L, Antonioli E, et al. Anaemia characterises patients with myelofibrosis harbouring Mpl mutation. Br J Haematol. 2007;137:244–7.
- Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med. 2013;369:2379–90.
- Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med. 2013;369:2391–405.
- Tefferi A, Lasho TL, Finke CM, Knudson RA, Ketterling R, Hanson CH, et al. CALR Vs JAK2 vs MPL-mutated or triple-negative myelofibrosis: clinical, cytogenetic and molecular comparisons. Leukemia 2014;28:1472–7.
- Chao MP, Gotlib J. Two faces of ET: CALR and JAK2. Blood 2014;123:1438–40.
- Tefferi A, Pardanani A. CALR Mutations and a new diagnostic algorithm for MPN. Nat Rev Clin Oncol. 2014;11:125–6.
- Li B, Xu J, Wang J, Gale RP, Xu Z, Cui Y, et al. Calreticulin mutations in Chinese with primary myelofibrosis. Haematologica 2014;99:1697–700.
- Tefferi A, Thiele J, Orazi A, Kvasnicka HM, Barbui T, Hanson CA, et al. Proposals and rationale for revision of the World Health Organization diagnostic criteria for polycythemia vera, essential thrombocythemia, and primary myelofibrosis: recommendations from an ad hoc international expert panel. Blood 2007;110:1092–7.
- Ruan GR, Chen SS, Li LD, Liu YR, Qin YZ, Li JL, et al. Detection of JAK2V617F mutation in patients with myeloproliferative disorders with TaqMan-MGB probe. Zhonghua Yi Xue Za Zhi. 2007;87:2401–4.
- Rumi E, Pietra D, Ferretti V, Klampfl T, Harutyunyan AS, Milosevic JD, et al. JAK2 Or CALR mutation status defines subtypes of essential thrombocythemia with substantially different clinical course and outcomes. Blood 2014;123:1544–51.
- Rotunno G, Mannarelli C, Guglielmelli P, Pacilli A, Pancrazzi A, Pieri L, et al. Impact of calreticulin mutations on clinical and hematological phenotype and outcome in essential thrombocythemia. Blood 2014;123:1552–5.
- Chi J, Nicolaou KA, Nicolaidou V, Koumas L, Mitsidou A, Pierides C, et al. Calreticulin gene exon 9 frameshift mutations in patients with thrombocytosis. Leukemia 2014;28:1152–4.
- Chi J, Manoloukos M, Pierides C, Nicolaidou V, Nicolaou K, Kleopa M, et al. Calreticulin mutations in myeloproliferative neoplasms and new methodology for their detection and monitoring. Ann Hematol. 2015;94:399–408.