It might seem strange for an article to focus largely on the history of a single mitochondrial haplogroup in an era when complete genome sequencing is becoming more common. But as recent publications and film documentaries have shown (Meldrum Citation2009; Oppenheimer et al. Citation2014; Smoot et al. Citation2010; Stanford and Bradley Citation2012), there is still considerable confusion about what the structure of mitochondrial genetic diversity in the Americas means for Native American population history. Specifically, there are persistent claims that the presence of mitochondrial haplogroup X2a in Native American populations is evidence for ancient trans-Atlantic gene flow from Europe or the Middle East into North America (Meldrum Citation2009; Oppenheimer et al. Citation2014; Smoot et al. Citation2010; Stanford and Bradley Citation2012). If true, this genetic evidence would lend considerable support to the Solutrean hypothesis, which suggests that the North American Clovis culture (13,300–12,800 cal yr BP) is directly descended from the Solutrean culture of southwestern Europe (23,500–18,000 cal yr BP). The current iteration of the Solutrean hypothesis was developed by Bruce Bradley and Dennis Stanford (Bradley and Stanford Citation2004; Stanford and Bradley Citation2012; see Abbott Citation1877, Greenman Citation1963 and Hibben Citation1941 for previous iterations of the hypothesis), and it has been heavily critiqued by many archaeologists (Eren et al. Citation2013, Citation2015; O'Brien et al. Citation2014; Philips Citation2014; Sellet Citation1998; Straus et al. Citation2005).
The idea that haplogroup X2a is derived from an ancient trans-Atlantic migration to the Americas has been repeatedly considered — and rejected — by anthropological geneticists over the last two decades (Brown et al. Citation1998; Fagundes et al. Citation2008; Reidla et al. Citation2003; Smith et al. Citation1999, 2005). However, we revisit it here because it continues to be discussed and because recently published genomic data from ancient and contemporary North Americans help clarify the population history of North America and the likely history of this haplogroup.
1. The current model
Several decades of studies analyzing classical genetic markers, DNA sequences from single loci, and, very recently, genome wide variants have cumulatively shown that all Native Americans derive ancestry from a fairly small founder population that likely occupied Beringia during the Last Glacial Maximum (LGM; ∼28,000 and 18,000 cal yr BP) (reviewed in Hoffecker et al. Citation2014; Kemp and Schurr Citation2010; Raff and Bolnick Citation2014). There is still a great deal that is unknown about this founder population, but current genetic evidence shows that it was descended from peoples in eastern Siberia who were related to the ancestors of both contemporary East Asians and contemporary West Eurasians (Kemp and Schurr Citation2010; O'Rourke and Raff Citation2010; Raghavan et al. Citation2014b). Some Native Americans also share ancestry with contemporary Austro-Melanesians, although it is debated whether these affinities reflect (a) gene flow from the Asian ancestors of Austro-Melanesians into Beringia before people moved into the Americas (Skoglund et al. Citation2015), or (b) gene flow after the founder population had entered and dispersed through the Americas (Raghavan et al. Citation2015). Either way, this founder population likely inhabited northeastern Siberia (Madsen Citation2015) or Beringia (Hoffecker et al. Citation2014) for an extended period of time (∼7000–15,000 years), possibly in isolated refugia (Hoffecker et al. Citation2014; Kitchen et al. Citation2008; Mulligan et al. Citation2008; Raghavan et al. Citation2015). During that time, a number of genetic variants evolved through mutation and genetic drift, and many of those variants are found only (or primarily) in the Americas today (Tamm et al. Citation2007).
The precise history of migration out of Siberia/Beringia following the LGM is currently the focus of active research and debate in anthropological genetics, archaeology, and geology. It is clear that people reached Monte Verde in Chile by 14,600 cal yr BP via a coastal route (Dillehay et al. Citation2008). By 13,000 cal yr BP, descendants of the Beringian founder population had differentiated into two major genetic groups (clades) in the Americas — one containing individuals from both North and South America (including the 12,600 cal yr BP Anzick-1 infant from Montana and the 8690–8400 cal yr BP Kennewick Man from Washington), and one restricted to North America (Raghavan et al. Citation2015). It has also been suggested based on mitochondrial DNA that some individuals from the ancestral Siberian/Beringian population moved into the Americas via a second interior route when an ice-free corridor opened between the Cordilleran and Laurentide ice sheets, after 13,500 cal yr BP (Perego et al. Citation2009). This migration might help account for the observed genetic structure in North and South America, but more genomic data from North American populations is needed to test this hypothesis (Achilli et al. Citation2013; Raghavan et al. Citation2015). Additional population movements from Siberia, including migrations at approximately 4000 cal yr BP and 1000–800 cal yr BP, also contributed to the contemporary gene pool in northern North America (Raff et al. Citation2015; Raghavan et al. Citation2014b; Reich et al. Citation2012). Other regions of the Americas saw more localized migration, gene flow, and genetic drift over the millennia, with the region-specific genetic structure of the Americas largely in place by 4000 cal yr BP (Raff et al. Citation2011). However, it is important to remember that the composition of these regional Native American gene pools may have been dramatically reshaped by post-1492 contact with Europeans, which brought warfare, disease, enslavement, removals and relocations, and extensive admixture (Bolnick et al. Citation2006; Crawford Citation1998; Dobyns Citation1983; Livi-Bacci Citation2006; Malhi et al. 2008; Reich et al. Citation2012; Smith Citation1987; Ubelaker Citation2006; Zlojutro et al. 2009). Thus, contemporary Native American populations are genetically diverse, deriving varying proportions of their ancestry from ancient Siberia/Beringia, Europe, Asia, Africa, and elsewhere (Bolnick et al. Citation2006; Hunley and Healy Citation2011; Moreno-Estrada et al. Citation2013; Reich et al. Citation2012; Smith et al. Citation2014; Verdu et al. Citation2014; Zegura et al. Citation2004).
A small number of scholars contend that in addition to having ancient Beringian ancestry, North American populations also derived some genetic ancestry from a trans-Atlantic migration in prehistoric times. Proponents of the Solutrean hypothesis suggest that a trans-Atlantic migration brought individuals inhabiting southwestern Europe between 23,500 and 18,000 cal yr BP to the Americas (Oppenheimer et al. Citation2014; Stanford and Bradley Citation2012), while others have proposed a trans-Atlantic migration of ancient Hebrews from the Middle East a few thousand years ago (Meldrum Citation2009; Smoot et al. Citation2010). For the sake of clarity and brevity, we focus our discussion here on the validity of the two main lines of evidence presented as supporting an ancient trans-Atlantic migration: the presence of mitochondrial haplogroup X2a on the North American continent and a signal of “West Eurasian ancestry” within Native American genomes.
2. Haplogroup X2a
X2a (and the related, rare haplogroup X2g) is a uniquely North American haplogroup, found at the highest frequencies in Great Lakes populations and at lower frequencies in the Plains and Pacific Northwest. It appears to be completely absent in populations from Central and South America (Perego et al. Citation2009). Its presence in pre-European contact skeletal remains confirms that it was not the result of post-1492 admixture (Bolnick and Smith Citation2007; Malhi and Smith Citation2002; Rasmussen et al. 2015).
However, unlike the other American mitochondrial haplogroups (A–D), which have clear parental haplotypes persisting in contemporary Siberian populations, there is no clear record of the evolutionary history of X2a in any population (Fernandes et al. Citation2012; Reidla et al. Citation2003). X2a's “grand-parental” haplogroup, X2, is found throughout, at low levels today throughout much of the world, including in the Near East (where X is more common and therefore thought to have initially evolved), South Caucasus, Europe, Siberia, Central Asia, and North Africa (Reidla et al. Citation2003). It is important to note that while the Altai people in southern Siberia exhibit X2 (Derenko et al. Citation2001), their lineages are not ancestral to those of North Americans, and the presence of X2 there today appears to be the result of recent gene flow from the west (Reidla et al. Citation2003).
Thus, the intermediate lineages linking X2 and X2a appear to have been lost in contemporary populations, or are so rare that they have not yet been well studied. We might expect to find them in ancient populations, but our temporal and spatial coverage of ancient populations is still quite sparse.
Despite — or perhaps because of — this gap in the phylogeographic record for haplogroup X2, the presence of X2a in North America has been cited as evidence for two different trans-Atlantic migrations before European contact. First, Meldrum (Citation2009) and Smoot et al. (Citation2010) suggested that X2a is the result of an ancient Hebrew migration from the Middle East to North America approximately 2500 cal yr BP. This hypothesis is undermined, though, by four key findings: X2a is not found in the Middle East, none of the X2 lineages present in the Middle East are immediately ancestral to X2a, the date of coalescence for X2a (14,200–17,000 cal yr BP) significantly precedes the hypothesized migration from the Middle East (Perego et al. Citation2009), and haplogroup X2a was present in North America far earlier than the hypothesized Hebrew migration, having been found in the 8690–8400 cal yr BP Kennewick Man remains from Washington state (Rasmussen et al. 2015). Thus, X2a does not provide any evidence for an ancient Hebrew migration from the Middle East to North America.
Second, Stanford and Bradley (Citation2012) and Oppenheimer et al. (Citation2014) have hypothesized that haplogroup X2a was brought to North America via a Pleistocene migration of Solutreans from western Europe. Stanford and Bradley (Citation2012) specifically cite the high frequency of haplogroup X2 in the Orkney Islands near Scotland (in 7.24% of the sampled individuals; Helgason et al. 2001) as supporting the trans-Atlantic migration of X2a. Madsen (Citation2015) also notes that it is interesting that “a modern population with one of the highest percentages of the X2 clade, higher even than Native American populations, is found in the Orkney Islands off the coast of Scotland” (Madsen Citation2015, 213). However, the X2 haplotypes found in the Orkney Islands was not ancestral to X2a, so this particular observation is irrelevant to the genetic prehistory of the Americas. We therefore focus instead on Stanford and Bradley's (Citation2012) and Oppenheimer et al.’s (Citation2014) other arguments in support of the idea that X2a derives from a Solutrean migration.
They base this hypothesis on two main lines of reasoning: (1) lineages ancestral to X2a have not been found in Siberia, in contrast to the other American haplogroups, and (2) the phylogeographic distribution of X2a in North America places “the oldest and deepest X2a branch ancestry” in northeast Canada, supporting an “ultimately eastern introduction of X2a and X2g to the Americas consistent with an additional trans-Atlantic migration suggested by archaeological evidence” (Stanford and Bradley Citation2012, 763).
First, it seems to us a very large leap to go from the observation that haplogroup X2a's ancestors have not been observed in Siberia to the conclusion that the Solutrean hypothesis “offers the only credible route-explanation for the unique, substantial presence of West Eurasian-derived X2g and X2a in the Great Lakes region of north-east America, and their antiquity” (Oppenheimer et al. Citation2014, 769). Although X2a's ancestor (X2) is thought to have evolved in the Near East (part of “West Eurasia”), characterizing X2a as a “West Eurasian” haplogroup is inaccurate because X2a is found only in people with indigenous North American ancestry. Associating X2a with “West Eurasia” is like saying “Solutreans evolved in Africa”: each statement refers to a location where the ancestral population is thought to have lived long ago, but that location is not relevant to the question under consideration. In particular, because X2 dispersed from the Near East and became widely distributed throughout the world, its descendent lineage X2a need not have evolved in the same place.
To differentiate between a Solutrean and Beringian source for X2a, one must look instead at the phylogeography of the most recent ancestors of X2a (). X2a'j is the clade that unites X2a and its nearest sister clade, X2j (Fernandes et al. Citation2012; Reidla et al. Citation2003). The geographic distribution of X2a'j haplotypes — especially those with some of the defining mutations for X2a (indicating that they belong to the lineage that led to X2a) — would be informative to this question, but no contemporary or ancient individuals belonging to these lineages have been identified, with the possible exception of one individual from Iran with the X2a'j defining transition at mitochondrial nucleotide position 12397. However, because this transition has been observed in other haplogroups and is known to occur recurrently, it is unclear if this Iranian individual belongs to the X2a'j lineage or not (Reidla et al. Citation2003). X2a's sister clade, X2j, is also extremely rare, being found in just a few contemporary individuals from Iran and Egypt (Fernandes et al. Citation2012). It is possible that the common ancestor of X2a and X2j originated there, but without identifying more individuals bearing X2j or X2a'j lineages, any inferences about the geographic origins of X2a'j or X2a are very tenuous.
Figure 1. Phylogenetic relationships of the X haplogroups mentioned in the text. Tree structure and diagnostic mutations from PhyloTree (van Oven and Kayser 2009). Dates (in cal yr BP) for the coalescence of haplogroup X2 and subclades are maximum likelihood estimates using the complete mitochondrial genome (Fagundes et al. 2012). Geographic distribution of clades from Fagundes et al. (2012) and Reidla et al. (2003). Note the considerable independent evolution that has occurred on the X2g, X2a, and X2j lineages. X2g’s mutational motif and phylogenetic position is noted to be “preliminary” and “likely to be further refined as additional sequences become available” on PhyloTree.
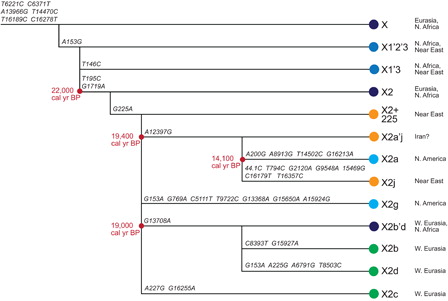
Thus, at this time, there is simply no evidence that X2a evolved in the Near East, Europe, or anywhere in West Eurasia. Stanford and Bradley (Citation2012) and Oppenheimer et al. (Citation2014) argue that the absence of evidence for X2a in West Eurasia is not evidence of absence, but of course, the same holds for Siberia. There is no compelling reason to think that X2a is more likely to have come from Europe than Siberia.
Where does this leave us? Until we have better geographic sampling of ancient DNA from the relevant time periods, the only way we can deduce anything about X2a's history is by studying it within North America or by making inferences based on the geographic distribution of other haplogroups or the patterns at other genetic loci. This indirect approach has been used in numerous studies, which have looked at the geographic distribution of X2a within North America to try to infer a migration route, compared the coalescence date of X2a to that of other founder haplogroups in the Americas, and examined the demographic histories of each haplogroup to see if they underwent similar evolutionary pressures (such as population bottlenecks and expansions) (Achilli et al. Citation2008; Fagundes et al. Citation2008; Kashani et al. Citation2012; Perego et al. Citation2009; Smith et al. Citation1999, 2005; Tamm et al. Citation2007). These studies have all reached the same conclusion and suggest that haplogroup X2a is likely to have originated in the same population(s) as the other American founder haplogroups, by virtue of having comparable coalescence dates and demographic histories.
Stanford and Bradley (Citation2012) and Oppenheimer et al. (Citation2014) have also argued in support of the Solutrean hypothesis by suggesting that the phylogeographic distribution of X2a in North America places the oldest (most basal) X2a lineages in northeastern Canada, which would be more consistent with an eastern introduction (and trans-Atlantic migration) of this haplogroup to the Americas. However, the newly published findings from the genome of Kennewick Man of Washington state are clearly at odds with this assessment. Kennewick Man's remains were directly dated to 8690–8400 cal yr BP, and the question of his affiliation with European (and Polynesian) populations was explicitly tested with genome-wide data — and rejected (Rasmussen et al. 2015). He is closely related to other ancient and contemporary Native Americans and shows clear Siberian affinities, with no recent European ancestry. Significantly, Kennewick Man's mitochondrial DNA belongs to haplogroup X2a, and he exhibits the most basal X2a lineage yet found (Rasmussen et al. 2015). Because this study places the oldest and most basal X2a lineage in the Pacific Northwest, it deals a serious blow to claims of genetic support for the Solutrean hypothesis. By Oppenheimer et al.'s own logic, this finding supports a Beringian, not Solutrean, origin for X2a. Thus, Kennewick Man's genome strongly suggests that X2a was not the result of trans-Atlantic gene flow.
3. “West Eurasian Ancestry” in Native Americans
What about evidence elsewhere in the genome for West Eurasian ancestry in Native Americans, which Oppenheimer et al. (Citation2014) also cite as supporting the Solutrean hypothesis? If the Solutrean hypothesis is correct, there ought to be ancient European ancestry in at least some Native American populations, reflecting the genetic contributions of the ancient Solutrean mariners who would have populated the East Coast. However, if their contribution to the Native American gene pool was small, it may only be detectable through high resolution analysis of Native American genomes — not through analyses of mitochondrial DNA alone. Accordingly, Oppenheimer et al. (Citation2014) have cited the 12,600 cal yr BP Anzick-1 genome from Montana (Rasmussen et al. Citation2014) and the 24,000 cal yr BP Mal'ta (MA-1) genome from south-central Siberia (Raghavan et al. Citation2014a) as showing “Pleistocene West Eurasian autosomal admixture in the Americas, which would be predicted by the SH [Solutrean Hypothesis]” (Oppenheimer et al. Citation2014, 766). Madsen (Citation2015, 228) has also suggested that “the genetic relationship of Native Americans to early west Eurasian populations makes it difficult to rule out a pre-glacial entry point in eastern North America.”
Raghavan et al. (Citation2014b) did estimate, based on the Mal'ta genome, that 14–38% of Native American ancestry is derived from a population ancestral to contemporary western Eurasians, and Rasmussen et al. (Citation2014) concluded that the Anzick-1 individual had ancestry from the same ancient population. However, it is important to parse these statements carefully, and we note that these studies did not find that the Pleistocene ancestors of Native Americans lived in West Eurasia. Rather, as Raghavan et al. (2014b) suggested, it appears that the relevant ancestral population lived farther to the east 24,000 years ago, extending into south-central Siberia, and the descendants of this ancient population contributed to both the contemporary West Eurasian gene pool and the Native American gene pool.
Furthermore, a suite of genomic studies has recently shown that the contemporary European gene pool emerged only in the last 8000 years or so, following large-scale migrations from the east and extensive admixture and population replacements in western Eurasia (Allentoft et al. Citation2015; Gamba et al. Citation2014; Haak et al. 2015; Lazaridis et al. Citation2014). We are unable to make direct genetic comparisons between Native Americans and the Solutrean peoples of southwestern Europe because no ancient DNA studies have included samples from Solutrean-associated skeletal remains, but the genomic data that are available from pre-Neolithic hunter–gatherers in Europe suggest that they were not closely related to Native Americans (Allentoft et al. Citation2015; Haak et al. 2015; Lazaridis et al. Citation2014). In other words, changes in the European gene pool over the last 8000 years have resulted in contemporary West Eurasians being more genetically similar to Native Americans than earlier Europeans were. Consequently, the genetic connections we see between Native Americans and West Eurasians today do not indicate connections between Native Americans and western Europeans in Pleistocene times.
Oppenheimer et al. (Citation2014) noted that few studies have formally tested the trans-Atlantic migration hypothesis alongside other models, and we agree that it is indeed important to do so. Rasmussen et al. (Citation2014) did evaluate the genetic affinities of the Anzick-1 individual with 143 contemporary populations in Eurasia, and determined that this ancient Native American was only distantly related to western Eurasian populations. Likewise, Rasmussen et al. (2015) explicitly assessed the degree of shared genetic history between Kennewick Man and contemporary Europeans, and found no evidence for European ancestry in Kennewick Man's genome. Perhaps the best test to date, though, comes from Lazaridis et al.’s (Citation2014) study, which used ancient and modern genomes to model the ancestral relationships between Eurasian, African, and Native American populations. The model that best fits their data was one in which the population ancestral to Native Americans was derived from ancient North Eurasian and East Asian sources, while contemporary Europeans were derived from ancient North Eurasian and West Eurasian sources (Lazaridis et al. Citation2014). In other words, gene flow was from the ancestral North Eurasian population into both the ancestral Native American and ancestral European populations. Lazaridis et al. (Citation2014) did not find any evidence of Pleistocene gene flow directly from West Eurasians into Native Americans. Their model is also consistent with other studies, which have shown that 62–86% of Native American ancestry derives from East Asia (Raghavan et al. Citation2014b).
Altogether, these results seem more consistent with migration to the Americas through East Asia and Siberia, as they are most parsimoniously explained by admixture between groups in Siberia (rather than in the Americas following separate migrations through Beringia and across the Atlantic). If West Eurasian alleles instead reached the Americas via a Solutrean migration after the initial movement into the Americas from Beringia, as Oppenheimer et al. (Citation2014) suggest, we would not expect to see their signature present uniformly across all Native American populations, but rather distributed in a gradient, with the highest levels around the presumed landing point of the Solutreans. Instead, the signature of West Eurasian ancestry is found equally in all Native American genomes tested to date, and therefore predates the evolution of regional genetic structure within North and South America (Raghavan et al. Citation2014a).
4. Concluding thoughts
We remain unconvinced by the arguments advanced thus far in favor of a trans-Atlantic migration prior to 1500 cal yr BP or so. As we have discussed, X2a has not been found anywhere in Eurasia, and phylogeography gives us no compelling reason to think it is more likely to come from Europe than from Siberia. Furthermore, analysis of the complete genome of Kennewick Man, who belongs to the most basal lineage of X2a yet identified, gives no indication of recent European ancestry and moves the location of the deepest branch of X2a to the West Coast, consistent with X2a belonging to the same ancestral population as the other founder mitochondrial haplogroups. Nor have any high-resolution studies of genome-wide data from Native American populations yielded any evidence of Pleistocene European ancestry or trans-Atlantic gene flow.
It is of course possible that genetic evidence of an ancient trans-Atlantic migration event simply has not been found yet. Should credible evidence of direct gene flow from an ancient Solutrean (or Middle Eastern) population be found within ancient Native American genomes, it would require the field to reassess the “Beringian only” model of prehistoric Native American migration. However, no such evidence has been found, and the Beringian migration model remains the best interpretation of the genetic, archaeological, and paleoclimate data to date.
Acknowledgements
We thank Dennis O'Rourke, Metin Eren, Michael Waters, Kelly Graf, Rick Smith, Lauren Springs, and Aida Miró-Herrans for helpful comments on earlier versions of this manuscript.
Additional information
Notes on contributors
Jennifer A. Raff
Jennifer Raff is an Assistant Professor in the Department of Anthropology at the University of Kansas. She trained in a dual program in Genetics, Molecular, Cell, Developmental Biology and Biological Anthropology at Indiana University, and conducted subsequent postdoctoral research at the University of Utah, Northwestern University, and the University of Texas. Her research focuses on the genetics of Native Americans in both ancient and contemporary populations, and encompasses questions about the initial peopling of the Americas, as well as region-specific histories in the North American Arctic and Midcontinental United States. She is also actively engaged in public outreach on issues pertaining to physical anthropology, genetics, and scientific literacy through writing (primarily at www.violentmetaphors.com), public speaking, and social media (primarily at https://twitter.com/JenniferRaff).
Deborah A. Bolnick
Deborah Bolnick is an Associate Professor of Anthropology and affiliated with the Population Research Center at the University of Texas at Austin. She received her Ph.D. in Anthropology from the University of California at Davis. Her research examines genetic variation in Native American populations and how it has been shaped by culture, history, and geography. As part of this research, she collaborates with indigenous individuals in the southern US, and analyzes DNA from both ancient and contemporary populations to track changes in genetic diversity over time and to help reconstruct population history in the Americas. She is also interested in the ethical, legal, social, and political implications of genetic research, including the ways in which genetic ancestry studies intersect with Euro-American and indigenous ideas about race, ethnicity, and identity.
References
- Abbott, C. C. 1877. “On the discovery of supposed paleolithic implements from the glacial drift in the valley of the Delaware River, Near Trenton, New Jersey.” Annual Report of the Peabody Museum of American Archaeology and Ethnology 10: 30–43.
- Achilli, A., U. A. Perego, C. M. Bravi, M. D. Coble, Q.-P. Kong, S. R. Woodward, A. Salas, A. Torroni, and H.-J. Bandelt. 2008. “The phylogeny of the four pan-American mtDNA haplogroups: Implications for evolutionary and disease studies.” PLoS One 3(3): e1764.
- Achilli, A., U. A. Perego, H. Lancioni, A. Olivieri, F. Gandini, B. H. Kashani, V. Battaglia, V. Grugni, N. Angerhofer, M. P. Rogers, R. J. Herrera, S. R. Woodward, D. Labuda, D. G. Smith, J. S. Cybulski, O. Semino, R. S. Malhi, and A. Torroni. 2013. “Reconciling migration models to the Americas with the variation of North American native mitogenomes.” Proceedings of the National Academy of the Sciences USA 110: 14308–14313.
- Allentoft, M. E., M. Sikora, K. G. Sjögren, S. Rasmussen, M. Rasmussen, J. Stenderup, P. B. Damgaard, H. Schroeder, T. Ahlström, L. Vinner, A. S. Malaspinas, A. Margaryan, T. Higham, D. Chivall, N. Lynnerup, L. Harvig, J. Baron, P. Della Casa, P. Dąbrowski, P. R. Duffy, A. V. Ebel, A. Epimakhov, K. Frei, M. Furmanek, T. Gralak, A. Gromov, S. Gronkiewicz, G. Grupe, T. Hajdu, R. Jarysz, V. Khartanovich, A. Khokhlov, V. Kiss, J. Kolář, A. Kriiska, I. Lasak, C. Longhi, G. McGlynn, A. Merkevicius, I. Merkyte, M. Metspalu, R. Mkrtchyan, V. Moiseyev, L. Paja, G. Pálfi, D. Pokutta, Ł. Pospieszny, T. D. Price, L. Saag, M. Sablin, N. Shishlina, V. Smrčka, V. I. Soenov, V. Szeverényi, G. Tóth, S. V. Trifanova, L. Varul, M. Vicze, L. Yepiskoposyan, V. Zhitenev, L. Orlando, T. Sicheritz-Pontén, S. Brunak, R. Nielsen, K. Kristiansen, and E. Willerslev. 2015. “Population genomics of Bronze Age Eurasia.” Nature 522: 167–172.
- Bolnick, D. A., D. I. Bolnick, and D. G. Smith. 2006. “Asymmetric male and female genetic histories among native Americans from eastern North America.” Molecular Biology and Evolution 23: 2161–2174.
- Bolnick, D. A., and D. G. Smith. 2007. “Migration and social structure among the Hopewell: Evidence from ancient DNA.” American Antiquity 72: 627–644.
- Bradley, B., and D. Stanford. 2004. “The North Atlantic ice-edge corridor: A possible Palaeolithic route to the New World.” World Archaeology 36: 459–478.
- Brown, M. D., S. H. Hosseini, A. Torroni, H.-J. Bandelt, J. C. Allen, T. G. Schurr, R. Scozzari, F. Cruciani, and D. C. Wallace. 1998. “mtDNA Haplogroup X: An ancient link between Europe/Western Asia and North America?” American Journal of Human Genetics 63: 1852–1861.
- Crawford, M. H. 1998. The Origins of Native Americas: Evidence from Anthropological Genetics. Cambridge: Cambridge University Press.
- Derenko, M. V., T. Grzybowski, B. A. Malyarchuk, J. Czarny, D. Miscicka-Sliwka, and I. A. Zakharov. 2001. “The presence of mitochondrial haplogroup X in Altaians from South Siberia.” American Journal of Human Genetics 67: 237–241.
- Dillehay, T. D., C. Ramirez, M. Pino, M. B. Collins, J. Rossen, and J. D. Pino-Navarro. 2008. “Monte Verde: Seaweed, food, medicine, and the peopling of South America.” Science 320: 784–786.
- Dobyns, H. F. 1983. Their Number Become Thinned: Native American Population Dynamics in Eastern North America. Knoxville: University of Tennessee Press.
- Eren, M. I., M. T. Boulanger, and M. J. O'Brien. 2015. “The Cinmar discovery and the proposed pre-Late Glacial Maximum occupation of North America.” Journal of Archaeological Science: Reports 2: 708–713.
- Eren, M. I., R. J. Patten, M. J. O'Brien, and D. J. Meltzer. 2013. “Refuting the technological cornerstone of the Ice-Age Atlantic crossing hypothesis.” Journal of Archaeological Science 40: 2934–2941.
- Fagundes, N. J. R., R. Kanitz, R. Eckert, A. C. S. Valls, M. R. Bogo, F. M. Salzano, D. G. Smith, W. A. Silva, M. A. Zago, A. K. Ribeiro-dos-Santos, S. E. B. Santos, M. L. Petzl-Erler, and S. L. Bonatto. 2008. “Mitochondrial population genomics supports a single pre-Clovis origin with a coastal route for the peopling of the Americas.” American Journal of Human Genetics 82: 583–592.
- Fernandes, V., F. Alshamali, M. Alves, M. D. Costa, J. B. Pereira, N. M. Silva, L. Cherni, N. Harich, V. Cerny, P. Soares, M. B. Richards, and L. Pereira. 2012. “The Arabian cradle: Mitochondrial relicts of the first steps along the southern route out of Africa.” American Journal of Human Genetics 90: 347–55.
- Gamba, C., E. R. Jones, M. D. Teasdale, R. L. McLaughlin, G. Gonzalez-Fortes, V. Mattiangeli, L. Domboróczki, I. Kővári, I. Pap, A. Anders, A. Whittle, J. Dani, P. Raczky, T. F. Higham, M. Hofreiter, D. G. Bradley, and R. Pinhasi. 2014. “Genome flux and stasis in a five millennium transect of European prehistory.” Nature Communications 5: 5257.
- Greenman, E. F. 1963. “The Upper Paleolithic and the New World.” Current Anthropology 4: 41–91.
- Haak, Wolfgang, Iosif Lazaridis, Nick Patterson, Nadin Rohland, Swapan Mallick, Bastien Llamas, Guido Brandt, Susanne Nordenfelt, Eadaoin Harney, Kristin Stewardson, Qiaomei Fu, Alissa Mittnik, Eszter Bánffy, Christos Economou, Michael Francken, Susanne Friederich, Rafael Garrido Pena, Fredrik Hallgren, Valery Khartanovich, Aleksandr Khokhlov, Michael Kunst, Pavel Kuznetsov, Harald Meller, Oleg Mochalov, Vayacheslav Moiseyev, Nicole Nicklisch,Sandra L. Pichler, Roberto Risch, Manuel A. Rojo Guerra, Christina Roth, Anna Szécsényi-Nagy, Joachim Wahl, Matthias Meyer, Johannes Krause, Dorcas Brown, David Anthony, Alan Cooper, Kurt Werner Alt, and David Reich. 2015. “Massive migration from the steppe was a source for Indo-European languages in Europe.” Nature 522 (7555): 207–211.
- Helgason, A., E. Hickey, S. Goodacre, V. Bosnes, K. Stefánsson, R. Ward, and B. Sykes. 2001. “mtDNA and the Islands of the North Atlantic: Estimating the Proportions of Norse and Gaelic Ancestry.” American Journal of Human Genetics 68: 723–37.
- Hibben, F. 1941. “Evidences of early occupation in Sandia Cave, New Mexico, and other sites in the Sandia-Manzano region.” Smithsonian Miscellaneous Collection 99(23).
- Hoffecker, J. F., S. A. Elias, and D. H. O'Rourke. 2014. “Out of Beringia?” Science 343: 979–980.
- Hunley, K. L., and M. E. Healy. 2011. “The impact of founder effects, gene flow, and European admixture on Native American genetic diversity.” American Journal of Physical Anthropology 146: 530–538.
- Kashani, H. B., U. A. Perego, A. Olivieri, N. Angerhofer, F. Gandini, V. Carossa, H. Lancioni, O. Semino, S. R. Woodward, A. Achilli, and A. Torroni. 2012. “Mitochondrial haplogroup C4c: A rare lineage entering America through the ice-free corridor?” American Journal of Physical Anthropology 147: 35–39.
- Kemp, B. M., and T. G. Schurr. 2010. “Ancient and modern genetic variation in the Americas.” In Human Variation in the Americas, edited by Benjamin M. Auerbach, 12–50. Center for Archaeological Investigations, Occasional Paper No. 38. Carbondale: Southern Illinois University Press.
- Kitchen, A. M., M. M. Miyamoto, and C. J. Mulligan. 2008. “A three-stage colonization model for the peopling of the Americas.” PLoS One 3(2): e1596.
- Lazaridis, I., N. Patterson, A. Mittnik, G. Renaud, S. Mallick, K. Kirsanow, P. H. Sudmant, J. G. Schraiber, S. Castellano, M. Lipson, B. Berger, C. Economou, R. Bollongino, Q. Fu, K. I. Bos, S. Nordenfelt, H. Li, C. de Filippo, K. Prüfer, S. Sawyer, C. Posth, W. Haak, F. Hallgren, E. Fornander, N. Rohland, D. Delsate, M. Francken, J. M. Guinet, J. Wahl, G. Ayodo, H. A. Babiker, G. Bailliet, E. Balanovska, O. Balanovsky, R. Barrantes, G. Bedoya, H. Ben-Ami, J. Bene, F. Berrada, C. M. Bravi, F. Brisighelli, G. B. Busby, F. Cali, M. Churnosov, D. E. Cole, D. Corach, L. Damba, G. van Driem, S. Dryomov, J. M. Dugoujon, S. A. Fedorova, I. Gallego, Romero, M. Gubina, M. Hammer, B. M. Henn, T. Hervig, U. Hodoglugil, A. R. Jha, S. Karachanak-Yankova, R. Khusainova, E. Khusnutdinova, R. Kittles, T. Kivisild, W. Klitz, V. Kučinskas, A. Kushniarevich, L. Laredj, S. Litvinov, T. Loukidis, R. W. Mahley, B. Melegh, E. Metspalu, J. Molina, J. Mountain, K. Näkkäläjärvi, D. Nesheva, T. Nyambo, L. Osipova, J. Parik, F. Platonov, O. Posukh, V. Romano, F. Rothhammer, I. Rudan, R. Ruizbakiev, H. Sahakyan, A. Sajantila, A. Salas, E. B. Starikovskaya, A. Tarekegn, D. Toncheva, S. Turdikulova, I. Uktveryte, O. Utevska, R. Vasquez, M. Villena, M. Voevoda, C. A. Winkler, L. Yepiskoposyan, P. Zalloua, T. Zemunik, A. Cooper, C. Capelli, M. G. Thomas, A. Ruiz-Linares, S. A. Tishkoff, L. Singh, K. Thangaraj, R. Villems, D. Comas, R. Sukernik, M. Metspalu, M. Meyer, E. E. Eichler, J. Burger, M. Slatkin, S. Pääbo, J. Kelso, D. Reich, and J. Krause. 2014. “Ancient human genomes suggest three ancestral populations for present-day Europeans.” Nature 513: 409–413.
- Livi-Bacci, M. 2006. “The depopulation of Hispanic America after the conquest.” Population and Development Review 32: 199–232.
- Madsen, D. B. 2015. “A framework for the initial occupation of the Americas.” PaleoAmerica 1: 217–250.
- Malhi, R. S., and D. G. Smith. 2002. “Brief communication: Haplogroup X confirmed in prehistoric North America.” American Journal of Physical Anthropology 118: 84–86.
- Malhi, R. S., Gonzalez-Oliver, A., Kemp, B. M., and Schroeder, K. B. “Distribution of Y chromosomes among native North Americans: a study of Athapaskan population history.” American Journal of Physical Anthropology 137 (2008): 412–424.
- Meldrum, R. L. 2009. Rediscovering the Book of Mormon Remnant through DNA. Honeoye Falls, NY: Digital Legend Press.
- Moreno-Estrada, A., S. Gravel, F. Zakharia, J. A. McCauley, J. K. Byrnes, C. R. Givnoux, P. A. Ortiz-Tello, R. J. Martineq, D. J. Hedges, R. W. Morris, C. Eng, K. Sandoval, S. Acevedo-Acevedo, P. J. Norman, Z. Layrisse, P. Parham, J. C. Martinez-Cruzado, E. G. Burchard, M. L. Cuccaro, E. R. Martin, and C. D. Bustamante. 2013. “Reconstructing the population genetic history of the Caribbean.” PLoS Genetics 9(11): e1003925.
- Mulligan, C. J., A. Kitchen, and M. M. Miyamoto. 2008. “Updated three-stage model for the peopling of the Americas.” PLoS One 3(9): e3199.
- O'Brien, M. J., M. T. Boulanger, M. Collard, B. Buchanan, L. Tarle, L. G. Straus, and M. I. Eren. 2014. “On thin ice: Problems with Stanford and Bradley's proposed Solutrean colonization of North America.” Antiquity 88: 606–624.
- Oppenheimer, S., B. Bradley, and D. Stanford. 2014. “Solutrean hypothesis: Genetics, the mammoth in the room.” World Archaeology 46: 752–774.
- O'Rourke, D. H., and J. A. Raff. 2010. “The human genetic history of the Americas: The final frontier.” Current Biology 20: R202–R207.
- Perego, U. A., A. Achilli, N. Angerhofer, M. Accetturo, M. Pala, A. Olivieri, B. H. Kashani, K. H. Ritchie, R. Scozzari, Q.-P. Kong, N. M. Myres, A. Salas, O. Semino, H.-J. Bandelt, S. R. Woodward, and A. Torroni. 2009. “Distinctive Paleo-Indian migration routes from Beringia marked by two rare mtDNA haplogroups.” Current Biology 19: 1–8.
- Philips, K. 2014. “Solutrean seal hunters? Modeling trans-Atlantic migration parameters fundamental to the ‘Solutrean solution’ for the peopling of the Americas.” Journal of Anthropological Research 70: 573–600.
- Raff, J. A., and D. A. Bolnick. 2014. “Palaeogenomics: Genetic roots of the first Americans.” Nature 506: 162–163.
- Raff, J. A., D. A. Bolnick, J. Tackney, and D. H. O'Rourke. 2011. “Ancient DNA perspectives on American colonization and population history.” American Journal of Physical Anthropology 146: 503–514.
- Raff, J. A., M. Rzhetskaya, J. Tackney, and M. G. Hayes. 2015. “Mitochondrial diversity of Inupiat people from the Alaskan North Slope provides evidence for the origins of the Paleo- and Neo-Eskimo peoples.” American Journal of Physical Anthropology 157: 603–614.
- Raghavan, M., M. DeGiorgio, A. Albrechtsen, I. Moltke, P. Skoglund, T. S. Korneliussen, B. Grønnow, M. Appelt, H. C. Gulløv, T. M. Friesen, W. Fitzhugh, H. Malmström, S. Rasmussen, J. Olsen, L. Melchior, B. T. Fuller, S. M. Fahrni, T. Stafford Jr., V. Grimes, M. A. Renouf, J. Cybulski, N. Lynnerup, M. M. Lahr, K. Britton, R. Knecht, J. Arneborg, M. Metspalu, O. E. Cornejo, A. S. Malaspinas, Y. Wang, M. Rasmussen, V. Raghavan, T. V. Hansen, E. Khusnutdinova, T. Pierre, K. Dneprovsky, C. Andreasen, H. Lange, M. G. Hayes, J. Coltrain, V. A. Spitsyn, A. Götherström, L. Orlando, T. Kivisild, R. Villems, M. H. Crawford, F. C. Nielsen, J. Dissing, J. Heinemeier, M. Meldgaard, C. Bustamante, D. H. O'Rourke, M. Jakobsson, M. T. Gilbert, R. Nielsen, and E. Willerslev. 2014a. “The genetic prehistory of the New World Arctic.” Science 345: 1255832.
- Raghavan, M., P. Skoglund, K. E. Graf, M. Metspalu, A. Albrechtsen, I. Moltke, S. Rasmussen, T. W. Stafford Jr., L. Orlando, E. Metspalu, M. Karmin, K. Tambets, S. Rootsi, R. Mägi, P. F. Campos, E. Balanovska, O. Balanovsky, E. Khusnutdinova, S. Litvinov, L. P. Osipova, S. A. Fedorova, M. I. Voevoda, M. DeGiorgio, T. Sicheritz-Ponten, S. Brunak, S. Demeshchenko, T. Kivisild, R. Villems, R. Nielsen, M. Jakobsson, and E. Willerslev. 2014b. “Upper Paleolithic Siberian genome reveals dual ancestry of Native Americans.” Nature 505: 87–91.
- Raghavan, M., M. Steinrücken, K. Harris, S. Schiffels, S. Rasmussen, M. DeGiorgio, A. Albrechtsen, C. Valdiosera, M. C. Ávila-Arcos, A. S. Malaspinas, A. Eriksson, I. Moltke, M. Metspalu, J. R. Homburger, J. Wall, O. E. Cornejo, J. V. Moreno-Mayar, T. S. Korneliussen, T. Pierre, M. Rasmussen, P. F. Campos, P. de Baros Damgaard, M. E. Allentoft, J. Lindo, E. Metspalu, R. Rodríguez-Varela, J. Mansilla, C. Henrickson, A. Seguin-Orlando, H. Malmström, T. Stafford, Jr., S. S. Shringarpure, A. Moreno-Estrada, M. Karmin, K. Tambets, A. Bergström, Y. Xue, V. Warmuth, A. D. Friend, J. Singarayer, P. Valdes, F. Balloux, I. Leboreiro, J. L. Vera, H. Rangel-Villalobos, D. Pettener, D. Luiselli, L. G. Davis, E. Heyer, C. P. Zollikofer, M. S. Ponce de León, C. I. Smith, V. Grimes, K. A. Pike, M. Deal, B. T. Fuller, B. Arriaza, V. Standen, M. F. Luz, F. Ricaut, N. Guidon, L. Osipova, M. I. Voevoda, O. L. Posukh, O. Balanovsky, M. Lavryashina, Y. Bogunov, E. Khusnutdinova, M. Gubina, E. Balanovska, S. Fedorova, S. Litvinov, B. Malyarchuk, M. Derenko, M. J. Mosher, D. Archer, J. Cybulski, B. Petzelt, J. Mitchell, R. Worl, P. J. Norman, P. Parham, B. M. Kemp, T. Kivisild, C. Tyler-Smith, M. S. Sandhu, M. Crawford, R. Villems, D. G. Smith, M. R. Waters, T. Goebel, J. R. Johnson, R. S. Malhi, M. Jakobsson, D. J. Meltzer, A. Manica, R. Durbin, C. D. Bustamante, Y. S. Song, R. Nielsen, and E. Willerslev. 2015. “Genomic evidence for the Pleistocene and recent population history of Native Americans.” Science 349(6250): aab3884.
- Rasmussen, M., S. L. Anzick, M. R. Waters, P. Skoglund, M. DeGiorgio, T. W. Stafford Jr., S. Rasmussen, I. Moltke, A. Albrechtsen, S. M. Doyle, G. D. Poznik, V. Gudmundsdottir, R. Yadav, A-S. Malaspinas, S. S. V. White, M. E. Allentoft, O. E. Cornejo, K. Tambets, A. Eriksson, P. D. Heintzman, M. Karmin, T. S. Korneliussen, D. J. Meltzer, T. L. Pierre, J. Stenderup, L. Saag, V. M. Warmuth, M. C. Lopes, R. S. Malhi, S. Brunak, T. Sicheritz-Ponten, I. Barnes, M. Collins, L. Orlando, F. Balloux, A. Manica, R. Gupta, M. Metspalu, C. D. Bustamante, M. Jakobsson, R. Nielsen, and E. Willerslev. 2014. “The genome of a late Pleistocene human from a Clovis burial site in western Montana.” Nature 506: 225–229.
- M. Rasmussen, M. Sikora, A. Albrechtsen, T. S. Korneliussen, J. V. Moreno-Mayar, G. D. Poznik, C. P. Zollikofer, M. S. Ponce de León, M. E. Allentoft, I. Moltke, H. Jónsson, C. Valdiosera, R. S. Malhi, L. Orlando, C. D. Bustamante, T. W. Stafford Jr., D. J. Meltzer, R. Nielsen, E. Willerslev. “The ancestry and affiliations of Kennewick Man.” Nature (2015).
- Reich, D., N. Patterson, D. Campbell, A. Tandon, S. Mazieres, N. Ray, M. V. Parra, W. Rojas, C. Duque, N. Mesa, L. F. García, O. Triana, S. Blair, A. Maestre, J. C. Dib, C. M. Bravi, G. Bailliet, D. Corach, T. Hünemeier, M. C. Bortolini, F. M. Salzano, M. L. Petzl-Erler, V. Acuña-Alonzo, C. Aguilar-Salinas, S. Canizales-Quinteros, T. Tusié-Luna, L. Riba, M. Rodríguez-Cruz, M. Lopez-Alarcón, R. Coral-Vazquez, T. Canto-Cetina, I. Silva-Zolezzi, J. C. Fernandez-Lopez, A. V. Contreras, G. Jimenez-Sanchez, M. J. Gómez-Vázquez, J. Molina, A. Carracedo, A. Salas, C. Gallo, G. Poletti, D. B. Witonsky, G. Alkorta-Aranburu, R. I. Sukernik, L. Osipova, S. A. Fedorova, R. Vasquez, M. Villena, C. Moreau, R. Barrantes, D. Pauls, L. Excoffier, G. Bedoya, F. Rothhammer, J. M. Dugoujon, G. Larrouy, W. Klitz, D. Labuda, J. Kidd, K. Kidd, A. Di Rienzo, N. B. Freimer, A. L. Price, and A. Ruiz-Linares. 2012. “Reconstructing Native American population history.” Nature 488: 370–374.
- Reidla, M., T. Kivisild, E. Metspalu, K. Kaldma, K. Tambets, H. V. Tolk, J. Parik, E. L. Loogväli, M. Derenko, B. Malyarchuk, M. Bermisheva, S. Zhadanov, E. Pennarun, M. Gubina, M. Golubenko, L. Damba, S. Fedorova, V. Gusar, E. Grechanina, I. Mikerezi, J. P. Moisan, A. Chaventré, E. Khusnutdinova, L. Osipova, V. Stepanov, M. Voevoda, A. Achilli, C. Rengo, O. Rickards, G. F. De Stefano, S. Papiha, L. Beckman, B. Janicijevic, P. Rudan, N. Anagnou, E. Michalodimitrakis, S. Koziel, E. Usanga, T. Geberhiwot, C. Herrnstadt, N. Howell, A. Torroni, and R. Villems. 2003. “Origin and diffusion of mtDNA Haplogroup X.” American Journal of Human Genetics 73: 1178–1190.
- Sellet, F. 1998. “The French connection: Investigating a possible Clovis-Solutrean Link.” Current Research in the Pleistocene 15: 67–68.
- Skoglund, P., S. Mallick, M. C. Bortolini, N. Chennagiri, T. Hunemeler, M. L. Petzl-Erler, F. M. Salzano, N. Patterson, and D. Reich. 2015. “Genetic evidence for two founding populations of the Americas.” Nature 525: 104–108.
- Smith, M. T. 1987. Archaeology of Aboriginal Culture Change in the Interior Southeast: Depopulation during the Early Historic Period. Gainsville: University of Florida Press.
- Smith, D. G., R. S. Malhi, J. Eshleman, J. G. Lorenz, and F. A. Kaestle. 1999. “Distribution of mtDNA haplogroup X among native North Americans.” American Journal of Physical Anthropology 110: 271–284.
- Smith, R. W. A., K. Manriquez, J. Mata-Miguez, D. G. Smith, and D. A. Bolnick. 2014. “Genetic variation among Native North Americans with complex ancestries: Implications for contemporary tribal diversity and recent population history.” Paper presented at the American Association of Physical Anthropology Annual Meeting, Calgary, Alberta, April 9–12.
- Smith, David Glenn, Ripan S. Malhi, Jason A. Eshleman, Frederika A. Kaestle, and Brian M. Kemp. 2005. “Mitochondrial DNA Haplogroups of Paleoamericans in North America. In Paleoamerican Origins: Beyond Clovis”, edited by Robson Bonnichsen, Bradley T. Lepper, Dennis Stanford, and Michael R. Waters, pp. 243–254. College Station: Texas A&M University Press.
- Smoot, S., R. Stout, and B. McLerran. 2010. The Lost Civilizations of North America. DVD. Directed by R. Stout. Bountiful, UT: Digital Legends.
- Stanford, D. J., and B. Bradley. 2012. Across Atlantic Ice: The Origin of America's Clovis Culture. Berkeley: University of California Press.
- Straus, L. G., D. J. Meltzer, and T. Goebel. 2005. “Ice age Atlantis? Exploring the Solutrean-Clovis ‘connection’.” World Archaeology 37: 507–532.
- Tamm, E., T. Kivisild, M. Reidla, M. Metspalu, D. G. Smith, C. J. Mulligan, C. M. Bravi, O. Rickards, C. Martinez-Labarga, E. K. Khusnutdinova, S. A. Fedorova, M. V. Golubenko, V. A. Stepanov, M. A. Gubina, S. I. Zhadanov, L. P. Osipova, L. Damba, M. I. Voevoda, J. E. Dipierri, R. Villems, and R. S. Malhi. 2007. “Beringian standstill and spread of Native American founders.” PLoS One 2(9): e829.
- Ubelaker, D. H. 2006. “Population size: Contact to nadir.” In Handbook of North American Indians: Environment, Origins, and Population, edited by D. Stanford, B. D. Smith, D. H. Ubelaker, and E. J. E. Szathmary, 694–701. Washington, DC: Smithsonian Institution Press.
- van Oven and Kayser. 2009. “Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation.” Human Mutation 30(2): E386–E394. doi:10.1002/humu.20921.
- Verdu, P., T. J. Pemberton, R. Laurent, B. M. Kemp, A. Gonzalez-Oliver, C. Gorodezky, C. E. Hughes, M. R. Shattuck, B. Petzelt, J. Mitchell, H. Harry, T. William, R. Worl, J. S. Cybulski, N. A. Rosenberg, and R. S. Malhi. 2014. “Patterns of admixture and population structure in Native populations of northwest North America.” PLoS Genetics 10(8): e1004530.
- Zegura, S. L., T. M. Karafet, L. A. Zhivotovsky, and M. F. Hammer. 2004. “High-resolution SNPs and microsatellite haplotypes point to a single, recent entry of Native American Y chromosomes into the Americas.” Molecular Biology and Evolution 21: 164–175.
- Zlojutro M., Rubicz, R., Crawford, M.H. 2009. “Mitochondrial DNA and Y-chromosome variation in five eastern Aleut communities: evidence for genetic substructure in the Aleut population.” Annals of Human Biology 36: 511–526.
