Abstract
Subcutaneous delivery of concentrated monoclonal antibodies (mAbs) is complicated by the propensity of mAbs to self-associate at elevated concentrations, which can lead to undesirable solution properties such as aggregation and abnormally high viscosity. Therefore, the selection of mAb candidates with low propensity to self-associate during early antibody discovery can significantly reduce challenges that may occur later during antibody development. However, it is difficult to use conventional biophysical methods for measuring weak mAb self-interactions during antibody discovery given the large number of antibody candidates as well as their low concentrations and purities. Nevertheless, significant progress has been made recently in adapting conventional biophysical methods as well as developing new ones for early identification of mAbs with low self-association propensities, which we highlight in this editorial. These advances should improve the selection of mAb candidates suitable for the extreme requirements of concentrated formulations necessary for subcutaneous delivery of therapeutic antibodies.
1. Introduction
The widespread interest in using monoclonal antibodies (mAbs) as therapeutics is based on the relative ease of identifying antibody variants with high binding affinity and exquisite binding specificity, as well as the long circulation times and low toxicities of antibodies. Antibodies are also attractive therapeutics because similar expression and purification processes can be used to generate large amounts of different mAbs. These favorable attributes have led to several successful antibody drugs for treating diverse human disorders ranging from rheumatoid arthritis to cancer Citation[1]. The remarkable investment by the biopharmaceutical industry in the area of antibody therapeutics – as evidenced by the large number of mAbs currently in clinical trials Citation[2] – is a testament to the therapeutic potential of antibodies and their significant future impact on human health.
A particularly important attribute of antibodies such as IgGs is that they typically have excellent biophysical properties, including high solubility and folding stability. These properties have led to the widespread use of antibodies as diagnostic reagents, which are commonly formulated at dilute concentrations (≤ 1 mg/ml) and stored for years without significant loss of activity. Nevertheless, these dilute concentrations are typically too low for antibody therapeutics, especially for those administered via subcutaneous delivery (the preferred delivery route) Citation[3]. To achieve typical therapeutic dosages of tens of mgs of antibody per kg of body weight, large amounts of antibody (> 100 mg) must be delivered subcutaneously in relatively small volumes (< 2 ml), resulting in the need for concentrated antibody formulations (> 50 mg/ml).
The attractive and relatively consistent solution properties of different mAbs at dilute concentrations are poor indicators of their solution properties at high antibody concentrations. Many previous studies have established that mAbs display variable and difficult-to-predict solution properties at elevated mAb concentrations (e.g., Citation[4-7]). Some mAbs are prone to aggregate when concentrated, which is especially concerning because aggregates may lead to adverse immune responses and/or loss of therapeutic activity due to stimulation of neutralizing antibodies Citation[8]. mAbs can also display abnormally high viscosity when concentrated, which can preclude their use in subcutaneous delivery due to problems related to forcing viscous solutions through small needles Citation[3].
Therefore, there is considerable interest in selecting mAbs with optimal biophysical properties early in the discovery process. Indeed, there appears to be a significant opportunity to identify mAbs with both high binding affinity and solubility because many (tens to thousands) mAbs are typically selected against the same target molecule. Nevertheless, it is much harder to screen for properties such as solubility or viscosity compared to binding affinity. Antibody binding assays such as ELISA can be easily conducted in a parallel format in 96- or 384-well plates for hundreds to thousands of samples using unpurified solutions of extremely dilute antibodies (< 1 µg/ml). In contrast, solubility or viscosity measurements are much lower throughput, require purified antibodies at much higher concentrations (1 – 100 mg/ml) and are difficult to obtain during early antibody discovery.
2. Measurements of mAb self-interactions
An attractive alternative to direct measurement of mAb properties such as solubility and viscosity is to measure mAb self-interactions that underlie these and other solution properties. This is particularly attractive because self-interaction measurements can – at least in principle – be measured at much lower antibody concentrations, including those relevant to early antibody selection (< 0.1 mg/ml). Static and dynamic light scattering are the gold standards for measuring protein self-association () Citation[9-11]. Unfortunately, these invaluable methods are not well suited for early antibody discovery (during which hundreds to thousands of antibodies are screened) because they require: i) purified mAbs; and ii) relatively high mAb concentrations (1 – 10 mg/ml). However, automated cell culture and purification systems can be employed to at least partially alleviate this bottleneck. Related methods, including osmometry Citation[12], require even higher mAb concentrations, which also makes them difficult to use during early antibody discovery.
Figure 1. Overview of assays for measuring monoclonal antibody (mAb) self-association. These assays include (A) static light scattering, (B) self-interaction chromatography and (C) affinity-capture self-interaction nanoparticle spectroscopy.
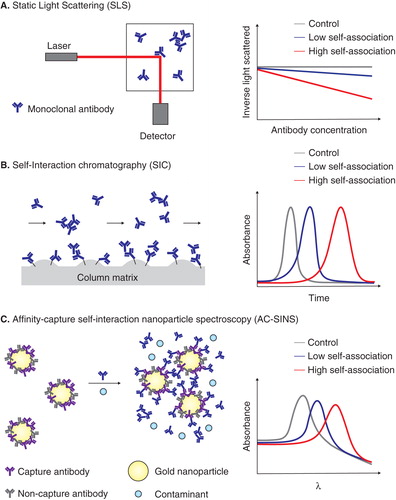
Several alternative methods for measuring mAb self-association have been developed that involve antibody immobilization on various types of surfaces. Traditional methods for measuring affinity interactions (e.g., Biacore or ELISA) use immobilized antibodies or antigens, but these methods are not sensitive enough to detect weak mAb self-interactions. One approach to improve the sensitivity of flow methods such as Biacore is to significantly increase the surface area for interaction, which is the basis of self-interaction chromatography (SIC; ) Citation[13,14]. In the latter approach, proteins such as mAbs are immobilized on highly porous chromatography particles in small columns, and the same proteins are passed through the corresponding columns. Attractive self-interactions lead to increased retention times, while the opposite is observed for repulsive interactions. The advantages of SIC are high sensitivity, the dilute mAb concentrations (as low as 0.1 – 1 mg/ml) required for each measurement, and the moderate throughput when using a chromatography system with an autosampler Citation[15-19]. Microfluidic versions of SIC have also been reported that further reduce the amount of mAb required for such measurements Citation[20,21]. However, the prime limitations of SIC are the preparation of a separate column for each mAb, and the fact that each measurement is made in series.
The utility of SIC has been improved by the surprisingly simple observation that immobilized mAbs can be replaced by non-specific polyclonal antibodies Citation[22]. Interestingly, this alternative form of SIC – referred to as cross-interaction chromatography (CIC) Citation[22-24] – is based on the premise that cross-interactions between mAbs and polyclonal antibodies are similar to mAb self-interactions. While exceptions to this simple hypothesis are expected, it is interesting that CIC is able to identify mAbs with both low and high propensity to self-associate, which has been verified via complementary assays Citation[18,22,25]. Nevertheless, it will be important in the future to evaluate whether CIC fails to identify some self-interactive antibodies with low polyspecificity. Moreover, while CIC dispenses with the high material consumption and time required for individual mAb immobilization, mAbs must still be purified and throughput is moderate.
Therefore, a particularly important challenge has been to develop higher throughput methods of measuring mAb self-interactions capable of processing hundreds of antibody samples in parallel. It would be most impactful if biophysical assays were developed that could measure weak mAb self-interactions in a 96- or 384-well plate format. Biolayer interferometry has recently been shown to be capable of measuring the weak interactions that typify poor solubility mAbs Citation[19]. This plate-based method requires little mAb (∼ 10 µg), but mAbs typically must be purified, concentrated to > 100 µg/ml and measured with specialized instruments.
Self-interaction nanoparticle spectroscopy (SINS) is another microplate assay that has been recently developed for characterizing mAb self-interactions () Citation[26-28]. The optical properties of gold colloid – including the wavelength of maximum absorbance (plasmon wavelength) – are dependent on the separation distance between particles. Thus, attractive self-interactions between mAbs immobilized on gold particles lead to reduced separation distances between gold particles and increased plasmon wavelengths (and vice versa for repulsive interactions). The utility of this assay has been significantly improved by first coating the gold particles with mixtures of polyclonal antibodies that bind human mAbs (i.e., capture antibodies) and non-specific antibodies that do not bind mAbs (i.e., non-capture antibodies) Citation[29]. This modified form of SINS – referred to as affinity-capture self-interaction nanoparticle spectroscopy or AC-SINS – enables the amount of immobilized mAb to be varied systematically by immobilizing different ratios of capture and non-capture antibodies. This leads to a significant improvement in assay sensitivity for discriminating between different mAbs. Other important attributes of AC-SINS are that mAbs can be captured from unpurified solutions, and that extremely low mAb concentrations (1 – 50 µg/ml) are required. Potential concerns with this approach are that antibody immobilization and/or non-specific interactions involving the gold surface can interfere with robust analysis of mAb self-association. Nevertheless, this high-throughput method has been used successfully to characterize a small set of mAbs, and awaits evaluation for use in antibody discovery.
3. Expert opinion
The significant push toward early assessment of the biophysical properties (e.g., solubility and viscosity) of therapeutic mAb candidates requires robust, high-throughput methods compatible with dilute and unpurified antibody samples. This need has stimulated a significant amount of research aimed at adapting conventional biophysical assays and developing new ones for measuring mAb self-association. The most promising assays are those that can measure mAb self-interactions for hundreds of mAbs in parallel and which are insensitive to impurities. To change the paradigm of antibody discovery, new biophysical assays need to be capable of similar throughput and robustness as ELISA assays, which are conducted for hundreds of samples in parallel using microtiter plates and do not require purified samples due to the use of capture antibodies. Thus, methods such as AC-SINS Citation[29] and related methods using interferometry Citation[19] appear to be most promising. Nevertheless, these methods have not been optimized for use during early antibody discovery and require additional method development. If such assays could be simplified to the point that they have similar sample and equipment requirements as ELISA assays, then they could be easily integrated into the antibody discovery process and provide unprecedented early insights into mAb properties that are indicative of high-concentration antibody behavior. This improved antibody selection process should greatly reduce the time and cost of antibody development by generating mAbs compatible with high-concentration formulation and delivery. Indeed, with such techniques, it may be possible to avoid difficult formulation and delivery problems by screening for better behaved mAbs early in the discovery process.
Declaration of interest
P Tessier has received research funding from the National Science Foundation (CBET grant 0954450), Merck, Eli Lilly, Genentech, MedImmune, and is a consultant for Adimab.
Bibliography
- Reichert JM. Marketed therapeutic antibodies compendium. mAbs 2012;4(3):413-15
- Reichert JM. Which are the antibodies to watch in 2013? mAbs 2013;5(1):1-4
- Daugherty AL, Mrsny RJ. Formulation and delivery issues for monoclonal antibody therapeutics. Adv Drug Deliv Rev 2006;58(5-6):686-706
- Yadav S, Liu J, Shire SJ, et al. Specific interactions in high concentration antibody solutions resulting in high viscosity. J Pharm Sci 2010;99(3):1152-68
- Kanai S, Liu J, Patapoff TW, Shire SJ. Reversible self-association of a concentrated monoclonal antibody solution mediated by Fab-Fab interaction that impacts solution viscosity. J Pharm Sci 2008;97(10):4219-27
- Dani B, Platz R, Tzannis ST. High concentration formulation feasibility of human immunoglubulin G for subcutaneous administration. J Pharm Sci 2007;96(6):1504-17
- Shire SJ, Shahrokh Z, Liu J. Challenges in the development of high protein concentration formulations. J Pharm Sci 2004;93(6):1390-402
- Ratanji KD, Derrick JP, Dearman RJ, et al. Immunogenicity of therapeutic proteins: influence of aggregation. J Immunotoxicol 2013. [Epub ahead of print]
- Scherer TM, Liu J, Shire SJ, et al. Intermolecular interactions of IgG1 monoclonal antibodies at high concentrations characterized by light scattering. J Phys Chem B 2010;114(40):12948-57
- Connolly BD, Petry C, Yadav S, et al. Weak interactions govern the viscosity of concentrated antibody solutions: high-throughput analysis using the diffusion interaction parameter. Biophys J 2012;103(1):69-78
- Sahin E, Grillo AO, Perkins MD, et al. Comparative effects of pH and ionic strength on protein-protein interactions, unfolding, and aggregation for IgG1 antibodies. J Pharm Sci 2010;99(12):4830-48
- Salinas BA, Sathish HA, Bishop SM, et al. Understanding and modulating opalescence and viscosity in a monoclonal antibody formulation. J Pharm Sci 2010;99(1):82-93
- Tessier PM, Lenhoff AM, Sandler SI. Rapid measurement of protein osmotic second virial coefficients by self-interaction chromatography. Biophys J 2002;82(3):1620-31
- Patro SY, Przybycien TM. Self-interaction chromatography: a tool for the study of protein-protein interactions in bioprocessing environments. Biotechnol Bioeng 1996;52(2):193-203
- Sule SV, Cheung JK, Antochshuk V, et al. Solution pH that minimizes self-association of three monoclonal antibodies is strongly dependent on ionic strength. Mol Pharm 2012;9(4):744-51
- Ahamed T, Esteban BN, Ottens M, et al. Phase behavior of an intact monoclonal antibody. Biophys J 2007;93(2):610-19
- Le Brun V, Friess W, Bassarab S, et al. A critical evaluation of self-interaction chromatography as a predictive tool for the assessment of protein-protein interactions in protein formulation development: a case study of a therapeutic monoclonal antibody. Eur J Pharm Biopharm 2010;75(1):16-25
- Bethea D, Wu SJ, Luo J, et al. Mechanisms of self-association of a human monoclonal antibody CNTO607. Protein Eng Des Sel 2012;25(10):531-7
- Sun T, Reid F, Liu Y, et al. High throughput detection of antibody self-interaction by bio-layer interferometry. mAbs 2013;5(6)
- Deshpande K, Ahamed T, van der Wielen LA, et al. Protein self-interaction chromatography on a microchip. Lab Chip 2009;9(4):600-5
- Garcia CD, Hadley DJ, Wilson WW, et al. Measuring protein interactions by microchip self-interaction chromatography. Biotechnol Prog 2003;19(3):1006-10
- Jacobs SA, Wu SJ, Feng Y, et al. Cross-interaction chromatography: a rapid method to identify highly soluble monoclonal antibody candidates. Pharm Res 2010;27(1):65-71
- Teske CA, Blanch HW, Prausnitz JM. Chromatographic measurement of interactions between unlike proteins. Fluid Phase Equilibria 2004;219(2):139-48
- Tessier PM, Sandler SI, Lenhoff AM. Direct measurement of protein osmotic second virial cross coefficients by cross-interaction chromatography. Protein Sci 2004;13(5):1379-90
- Spencer S, Bethea D, Raju TS, et al. Solubility evaluation of murine hybridoma antibodies. mAbs 2012;4(3):319-25
- Sule SV, Sukumar M, Weiss WF, et al. High-throughput analysis of concentration-dependent antibody self-association. Biophys J 2011;101(7):1749-57
- Bengali AN, Tessier PM. Biospecific protein immobilization for rapid analysis of weak protein interactions using self-interaction nanoparticle spectroscopy. Biotechnol Bioeng 2009;104(2):240-50
- Tessier PM, Jinkoji J, Cheng YC, et al. Self-interaction nanoparticle spectroscopy: a nanoparticle-based protein interaction assay. J Am Chem Soc 2008;130(10):3106-12
- Sule SV, Dickinson CD, Lu J, et al. Rapid analysis of antibody self-association in complex mixtures using immunogold conjugates. Mol Pharm 2013;10(4):1322-31
