Survival rates have not changed in the past 30 years.
*Percentage survival.
N: Lymph node staging; T: Tumor staging.
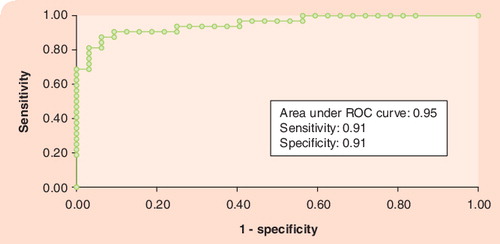
The final logistic model included four salivary mRNA biomarkers, interleukin (IL)-1B, ornithine decarboxylase antizyme-1, spermidine acetyltransferase and IL-8. Using a cut-off probability of 50%, we obtained sensitivity of 91% and specificity of 91% by ROC. The calculated area under the ROC curve was 0.95. ROC: Receiver operator characteristic.
Oral cancers annually affect 38,000 people in the USA and hundreds of thousands of others around the globe. Despite treatment advances, the disease’s overall 5-year survival rate has not improved in the past 30 years and remains among the worst of all cancers. One factor behind oral cancer’s high mortality is the challenge in its early detection. The use of saliva for the detection of oral cancer has been a historical goal that has yet to come to fruition. This review highlights translational research efforts, in alignment with initiatives sparked by the National Institute of Dental and Craniofacial Research (NIDCR), towards the realization of saliva diagnostics, particularly for saliva-based oral cancer detection.
The ability to monitor health status, disease onset and progression, and treatment outcome through noninvasive means is a most desirable goal in healthcare promotion and delivery. There are three prerequisites to materialize this goal: specific biomarkers associated with a health or disease state; a noninvasive approach to detect and monitor the biomarkers; and the technologies to discriminate the biomarkers. Oral fluid (saliva), which is the mirror of the body, is a perfect medium to be explored for health and disease surveillance.
A growing number of proof-of-principle examples have been established for the use of saliva to monitor systemic diseases and conditions. The barriers to widespread implementation of saliva diagnostics derive from technological problems, such as sensitivity, miniaturization, high throughput, automation, portability, low cost, high functionality and speed to enable detection and measurements of multiple disease markers in saliva have largely been overcome. Techniques are emerging from a combination of miniaturization technologies and discoveries in many different fields of biology, chemistry, physics and engineering are leading to high-throughput, automated, portable, low-cost, more efficient and rapid biochemical analyses. Miniaturized diagnostic technologies will be able to yield critical patient information with minute amounts of body fluids, thus reflecting health and disease status.
Oral cancer
Oral cancer is the sixth most common cancer in the USA, affecting 38,000 Americans annually and killing 7200. Worldwide, they annually affect between 350,000 Citation[101]. Over 90% of these cancers are squamous cell carcinoma. Despite treatment advances that have resulted in reductions in patient morbidity, the overall 5-year survival rate for oral squamous cell carcinoma (OSCC) remains among the worst of all cancer death rates (∼30–40% for the past few decades, which is considerably lower than those for colorectal, cervix and breast origin Citation[1,2]. One patient dies from oral cancer every hour in the USA. This high morbidity rate can be attributed to a number of factors, including nonresponsiveness to chemotherapy and radiation therapy, late presentation of lesions, and a lack of satisfactory biological markers for early lesion detection Citation[3]. According to The Oral Cancer Foundation, oral cancer is particularly dangerous because it has a high risk of developing second, primary tumors Citation[101]. Patients who survive a first encounter with the disease have up to a 20-fold increased risk of developing a second cancer.
Current oral cancer diagnostic & screening approaches
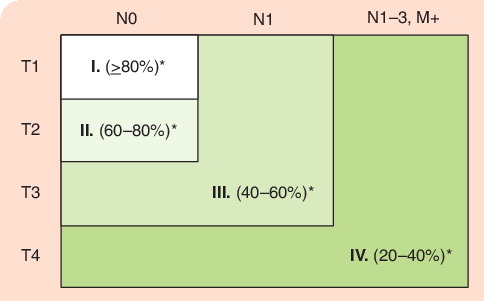
The most definitive procedure for oral cancer diagnosis is a scalpel biopsy followed by careful histopathological evaluation by a qualified pathologist. For this to be an effective procedure, it requires three consecutive events: a visit to the dentist and physician’s office, biopsy by the licensed healthcare provider and a pathologist’s evaluation. When effectively administered and reimbursed, as is in the Scandinavian countries, this will lead to early detection of oral cancer lesions that would otherwise have progress to later stage cancer, which carries a worse prognosis. illustrates the prognostic difference of an oral cancer lesion as it advances from an early to a late lesion. Detection of an oral cancer at Stage I will carry a prognosis of 80% survival, while the same lesion at Stage III carries a 20% survival. This is a dramatic difference that will affect not only the quality of life for the patient but will have significant savings on the healthcare costs on the medical treatments of a Stage I versus Stage III oral cancer patient.
With this in mind, scientists have been searching for alternative approaches to biopsy, with the hope to find a test for oral cancer detection that is similar to the Papanicolaou smear, which has significantly improved the mortality of cervical cancer. Since most oral cancers arise as asymptomatic small lesions, formal diagnosis procedures begin only when the clinician or patient notices abnormal tissues Citation[4]. Microscopic investigation of the progressive cancer is often conducted too late for successful intervention Citation[5]. It is also impractical to use imaging techniques for cancer screening, since they are time-consuming and expensive. These techniques are typically used for confirmation due to their insensitivity for small lesions Citation[6]. Studies have demonstrated that good positive-predictive values can be achieved by oral cancer tissue staining with toluidine blue Citation[7,8]. However, extensive experience is required in applying this technique and in interpreting its results. Exfoliative cytology may be a less invasive method for oral cancer detection Citation[9], but exfoliated cancer cells tend to correlate with tumor burden, and lower rates of detection are seen in those with minimal or early disease.
A number of molecular-based diagnostic markers have been used to detect the presence of OSCC with varying degrees of sensitivity and specificity. DNA markers include TP53, microsatellite instability, and presence of human papillomavirus (HPV) and Epstein–Barr virus genomic sequences Citation[10–13]. Cytokeratins have been used for RNA diagnostics, while squamous cell carcinoma, CD44, cytokeratin 19 fragments and telomerase have been used as protein markers Citation[14,15]. However, none of these markers universally identifies OSCC. Microsatellite markers are the most promising amongst these, where at least one of a panel of 23 markers can detect the presence of an oral cancer cell in the saliva of 79% of oral cancer patients Citation[16–19]. However, microsatellite instability analysis is not particularly sensitive and requires large amounts of cancer cell DNA, approximately one cancer cell among 200 normal cells. Furthermore, it is difficult to perform on a large number of clinical samples because many markers are necessary for testing Citation[16].
Recently, a number of clinical diagnostic aids for oral cancer detection have emerged. They include OralCDx® Brush Biopsy (CDx Laboratories, Inc.), ViziLite® Plus (Zila, Inc.) and toluidine blue. OralCDx Brush Biopsy is a noninvasive, chair-side procedure that determines whether an oral lesion is benign or potentially harmful. Precancerous and early-stage oral cancerous lesions can be determined. All suspicious lesions with abnormal cytology will be required for a biopsy follow-up. The ViziLite visual examination system is based on the differential densities of the nuclear content, and mitochondrial matrix of abnormal cells is typically greater than normal cells. The increased molecular density is believed to reveal the increased proliferative rate and metabolic activity of precancerous cells. The ViziLite exam enhances the examiner’s ability to see the difference in the nuclear/cytoplasmic ratio of dysplastic cells. After rinsing with a dilute acetic acid solution, the dense nucleus of abnormal squamous epithelium tissue will appear white when viewed under a diffuse low-energy wavelength light. Normal epithelium will absorb the light and appear dark. ViziLite can identify an abnormality, but a definitive diagnosis can only be made by biopsy.
History of using saliva for oral cancer molecular detection
The use of saliva for oral cancer screening or diagnostics is still in its infancy. Its use began following a report of a small study in Taiwan in 2000 by Liao and coworkers, which claimed that exon 4 codon 63 of the p53 gene is mutated in salivary DNA from five of eight (62.5%) oral cancer patients Citation[20]. Five of 27 control subjects (18.5%) had similar mutations in their p53 gene. In 2001, El-Naggar and coworkers demonstrated genetic heterogeneity in saliva from patients with OSCC and suggested the use of epithelial cells in saliva from patients with head and neck squamous tumorigenesis for genetic analysis Citation[21]. More recently, Jiang and coworkers reported the increase of mitochondrial DNA content in the saliva of head and neck cancer patients Citation[22]. Another report from the same group reported that quantitative analysis of HPV 16 DNA in salivary rinses enables detection of HPV-related head and neck cancer Citation[23]. However, the authors cautioned that specific limitations exist that prevent the application of this as a screening technique for a broad population.
University of California Los Angeles approach to saliva diagnostic for oral cancer
Our laboratory at the University of California Los Angeles (UCLA) utilizes research platforms towards the global identification of disease signatures in saliva. The premise of our approach is that serum contents, such as disease biomarkers, are largely present in saliva, thus rendering oral fluid a logical source to harness disease biomarkers Citation[24]. We employ both a proteome-wide as well as a genome-wide approach towards the identification of disease biomarkers and signatures.
Human salivary proteome as targets for human disease diagnostics
UCLA is one of the three NIDCR-funded groups to comprehensively decipher human salivary pathogenesis. We have already identified 309 distinct proteins in human whole saliva using 2D gel electrophoresis mass spectrometry (MS) and shotgun proteomics Citation[25]. This work was highlighted in the Journal of Proteome Research. Using a similar approach, we also conducted comparative proteome analysis of submandibular and sublingual saliva Citation[26].
To date, we have discovered two salivary proteins, interleukin (IL)-8 and thioredoxin, that can discriminate between saliva of oral cancer and control subjects. IL-8 was discovered through our previous tissue-based expression profiling effort Citation[27]. IL-8 is significantly elevated in saliva of oral cancer patients and is highly discriminatory of detecting oral cancer in saliva (n = 64) with an receiver operator characteristic (ROC) value of 0.95, sensitivity 86% and specificity 97% at a cut-off of 600 pg/ml Citation[27,28]. Of interest is that both IL-8 protein and RNA are concordantly increased for oral cancer patients Citation[28]. The concentrations of the IL-8 protein in saliva of oral cancer patients and control subjects are 750 ± 236 pg/ml and 250 ± 130 pg/ml, respectively. Similarly, oral cancer patients have significantly higher salivary IL-8 mRNA concentrations than control subjects. Due to the frequent inflammation association of this cytokine, we have further demonstrated that the oral cancer elevation of salivary IL-8 mRNA and protein is significantly higher than in advanced periodontitis patients.
These results enable us to conclude that, while severe inflammation in the oral cavity (e.g., in advanced periodontitis patients) does elevate salivary IL-8 protein and mRNA levels, it is not significant. Salivary IL-8 protein and mRNA levels in oral cancer patients are elevated significantly above those of control patients as well as advanced periodontitis patients, supporting the use of salivary IL-8 as a biomarker for oral cancer detection [Tan W and coworkers, Unpublished].
Thioredoxin was discovered as a salivary oral cancer biomarker by a proteomic approach using matrix-assisted laser desorption/ionization (MALDI) time-of-flight. We have established an integrated methodology to sequence candidate protein and/or peptide biomarkers. Using MALDI-MS profiling of saliva proteins, we have identified an approximately 11600-Da protein that was present at significantly higher levels in oral cancer saliva than in matched control subjects (p < 0.01). To identify this candidate biomarker, an oral cancer saliva sample was fractionated by reverse-phase liquid chromatography (LC; C4 column) followed by MALDI-MS of the LC fraction containing the candidate biomarker of approximately 11600 Da. This fraction was subsequently digested by trypsin for LC tandem MS (MS/MS) analysis. Mascot database searching indicated that four peptides were matched to this protein, with a sequence coverage of 31%. These results suggested that saliva thioredoxin is a validated biomarker for oral cancer detection [Hu S and coworkers, Unpublished].
Human salivary transcriptome as targets for cancer diagnostics
The UCLA research group has recently found that there are approximately 3000 human mRNAs in the cell-free saliva of normal subjects Citation[29]. Furthermore, there is a core signature of 185 mRNAs present in all normal subjects, which provides the rationale for the use the salivary transcriptome for disease diagnostics. The discovery that a large panel of human RNA can be reliably detected in saliva gives rise to the potential of this novel clinical approach. We evaluated the diagnostic value of this approach by using OSCC as the proof-of-principle disease, and found that seven salivary RNAs out of approximately 3000 mRNAs were consistently elevated in saliva from oral cancer patients. Of these, four in combination (ornithine decarboxylase antizyme-1, spermidine acetyltransferase, IL-8 and IL-1-β) have the ability to discriminate saliva from oral cancer patients from that of control subjects, with a ROC value of 0.95 a specificity of 91% and a sensitivity of 91% Citation[30]. While the initial study was performed on 64 subjects Citation[29], we have since carried out eight additional independent clinical detection studies on 350 subjects and found that the seven saliva mRNA biomarkers behaved very consistently with an overall accuracy rate of 85%. The discovery of RNA biomarkers in saliva that can have oral cancer discriminatory ability is a novel finding. This is now being explored for its translational potential and value.
It is currently under debate which bodily fluids (i.e., blood, saliva, urine or cerebral spinal fluid) are a more suitable clinically diagnostic for a specific disease entity. We have recently made that comparison for oral cancer. The same patients in which we identified saliva RNA signatures for oral cancer detection were examined for serum RNA signatures for oral cancer detection. Similar to saliva, four RNA biomarkers collectively can determine saliva of individuals with oral cancer, with a ROC value of 0.88 Citation[31]. While this is very good, the salivary RNA biomarkers have a ROC value of 0.95 Citation[30]. Thus, for oral cancer detection, saliva RNA biomarkers have a slight edge over serum RNA biomarkers.
Saliva diagnostics for other high-impact systemic diseases
Saliva has been examined for the detection of a number of systemic diseases ranging from infectious diseases Citation[32,33] (including HIV Citation[34,35]) to Alzheimer’s disease Citation[36]. Our laboratory has begun to explore a number of efforts to identify high-impact systemic diseases and explore their diagnostic signatures in saliva. Breast cancer is the first systemic disease to be explored for the presence of proteomic and genomic signatures in the saliva of breast cancer patients. It should be noted that Charlie Streckfus has reported that Her2 and CA15-3 levels are elevated in cancer versus control subjects’ saliva Citation[37,38]. It is also the intent of the UCLA group to perform rigorous proteome- and genome-wide discovery efforts to identify and validate salivary proteomic and genomic biomarkers in breast cancer patients. The research will follow guidelines from the National Cancer Institute Early Disease Research Network for biomarker validation Citation[39], similar to the ongoing oral cancer biomarker validation.
Future perspectives
While it is clear that there is a national agenda to turn saliva diagnostics into a clinical and commercial reality, much work is necessary before this vision can be realized. There remains the need to identify definitive disease-associated salivary biomarkers (proteins and genetic) that can be use in conjunction with the technology platforms for saliva diagnostics. The UCLA group is also poised to develop and validate the Oral Fluid NanoSensor Test as a point-of-care, chair-side, portable and multiplexible device to be used for saliva diagnostics . Collectively, technology platform advancement and the identification and validation of robust and discriminatory suites of salivary biomarkers for disease diagnostics represent the necessary marriage to propel saliva diagnostics into a clinical and commercial reality. At the same time, we are building the scientific foundation towards the use of saliva as a diagnostic fluid Citation[40]. The source of salivary biomarkers, proteins and RNA are currently being studied. Control mechanisms of salivary RNA turnover and the fate of these RNAs are also currently being address. This is a perfect example of translational research in reverse, based on a highly relevant clinical observation that saliva contains proteomic and genomic biomarkers for oral cancer detection, and building a scientific foundation towards the mechanistic background, thereby enabling better exploitation of the full clinical potential of saliva diagnostics.
Acknowledgements
David T Wong is supported by Public Health Service grants UO1 DE-15018, UO1 DE-16275, RO1 DE-15970 and the Jonsson Comprehensive Cancer Center.
References
- Schantz SP. Carcinogenesis, markers, staging, and prognosis of head and neck cancer. Curr. Opin. Oncol.5(3), 483–490 (1993).
- Schantz SP. Biologic markers, cellular differentiation, and metastatic head and neck cancer. Eur. Arch. Otorhinolaryngol.250(8), 424–428 (1993).
- Ferlito A, Shaha AR, Rinaldo A. The incidence of lymph node micrometastases in patients pathologically staged N0 in cancer of oral cavity and oropharynx. Oral Oncol.38(1), 3–5 (2002).
- Epstein JB, Zhang, L, Rosin, M. Advances in the diagnosis of oral premalignant and malignant lesions. J. Can. Dent. Assoc.68(10), 617–621 (2002).
- Fong K, Srivastava S, Gopal-Srivastava R, Kramer B et al. Molecular genetic basis for early cancer detection and cancer susceptibility. Mol. Pathol. Early Cancer13–26 (1999).
- Myers L, Wax M. Positron emission tomography in the evaluation of the negative neck in patients with oral cavity cancer. J. Otolaryngol.27(6), 342–347 (1998).
- Zhang L, Williams M, Poh CF et al. Toluidine blue staining identifies high-risk primary oral premalignant lesions with poor outcome. Cancer Res.65(17), 8017–8021 (2005).
- Mashberg A, Samit A. Early diagnosis of asymptomatic oral and oropharyngeal squamous cancers. CA Cancer J. Clin.45(6), 328–351 (1995).
- Rosin MP, Cheng X, Poh C et al. Use of allelic loss to predict malignant risk for low-grade oral epithelial dysplasia. Clin. Cancer Res.6(2), 357–362 (2000).
- Erber R, Conradt C, Homann N et al. TP53 DNA contact mutations are selectively associated with allelic loss and have a strong clinical impact in head and neck cancer. Oncogene16(13), 1671–1679 (1998).
- Gleich LL, Wang J, Gluckman JL, Fenoglio-Preiser CM. Microsatellite instability as a predictor of survival in head and neck cancer – is there a link with colon cancer? ORL J. Otorhinolaryngol. Relat. Spec.65(4), 193–198 (2003).
- Paz IB, Cook N, Odom-Maryon T, Xie Y, Wilczynski SP. Human papillomavirus (HPV) in head and neck cancer. An association of HPV 16 with squamous cell carcinoma of Waldeyer’s tonsillar ring. Cancer79(3), 595–604 (1997).
- Shimakage M, Horii K, Tempaku A, Kakudo K, Shirasaka T, Sasagawa T. Association of Epstein–Barr virus with oral cancers. Hum. Pathol.33(6), 608–614 (2002).
- Xu XC, Lee JS, Lippman SM, Ro JY, Hong WK, Lotan R. Increased expression of cytokeratins CK8 and CK19 is associated with head and neck carcinogenesis. Cancer Epidemiol. Biomarkers Prev.4(8), 871–876 (1995).
- Nagler R. Oral cancer – molecular aberrations and tumor markers cellular aberrations. Harefuah142(4), 272–276, 318, 317 (2003).
- Sidransky D. Emerging molecular markers of cancer. Nature Rev. Cancer3, 210–219 (2002).
- Ishwad CS, Ferrell RE, Rossie KM et al. Microsatellite instability in oral cancer. Int. J. Cancer64(5), 332–335 (1995).
- Nunes DN, Kowalski LP, Simpson AJ. Detection of oral and oropharyngeal cancer by microsatellite analysis in mouth washes and lesion brushings. Oral Oncol.36(6), 525–528 (2000).
- Shin KH, Park KH, Hong HJ et al. Prevalence of microsatellite instability, inactivation of mismatch repair genes, p53 mutation, and human papillomavirus infection in Korean oral cancer patients. Int. J. Oncol.21(2), 297–302 (2002).
- Liao PH, Chang YC, Huang MF, Tai KW, Chou MY. Mutation of p53 gene codon 63 in saliva as a molecular marker for oral squamous cell carcinomas. Oral Oncol.36(3), 272–276 (2000).
- El-Naggar AK, Mao L, Staerkel G et al. Genetic heterogeneity in saliva from patients with oral squamous carcinomas: implications in molecular diagnosis and screening. J. Mol. Diagn.3(4), 164–170 (2001).
- Jiang WW, Masayesva B, Zahurak M et al. Increased mitochondrial DNA content in saliva associated with head and neck cancer. Clin. Cancer Res.11(7), 2486–2491 (2005).
- Zhao M, Rosenbaum E, Carvalho AL et al. Feasibility of quantitative PCR-based saliva rinse screening of HPV for head and neck cancer. Int. J. Cancer117(4), 605–610 (2005).
- Phillips C. Rinse and spit: saliva as a cancer biomarker source. In: NCI Cancer Bulletin,3–5 (2006).
- Hu S, Xie Y, Ramachandran P et al. Large-scale identification of proteins in human salivary proteome by liquid chromatography/mass spectrometry and two-dimensional gel electrophoresis-mass spectrometry. Proteomics5(6), 1714–1728 (2005).
- Hu S, Denny P, Denny P et al. Differentially expressed protein markers in human submandibular and sublingual secretions. Int. J. Oncol.25(5), 1423–1430 (2004).
- Alevizos I, Mahadevappa M, Zhang X et al. Oral cancer in vivo gene expression profiling assisted by laser capture microdissection and microarray analysis. Oncogene20(43), 6196–6204 (2001).
- St John M, Li Y, Zhou X et al. IL-6 and IL-8: potential biomarkers for oral cavity and oropharyngeal SCCA. Arch. Otolaryngol. Head Neck Surg.130, 929–935 (2004).
- Li Y, Zhou X, St John MA, Wong DT. RNA profiling of cell-free saliva using microarray technology. J. Dent. Res.83(3), 199–203 (2004).
- Li Y, St John MA, Zhou X et al. Salivary transcriptome diagnostics for oral cancer detection. Clin. Cancer Res.10(24), 8442–8450 (2004).
- Li Y, Elashoff D, Oh M et al. Serum circulating human mRNA profiling and its utility for oral cancer detection. J. Clin. Oncol.24(11), 1754–1760 (2006).
- Parry J. Simple and reliable salivary tests for HIV and hepatitis A and B virus diagnosis and surveillance. Ann. NY Acad. Sci.694, 216–233 (1993).
- Nair P, Schroeder, H. Duct-associated lymphoid tissue (DALT) of minor salivary glands and mucosal immunity. Immunology57, 171–180 (1986).
- Malamud D. Oral diagnostic testing for detecting human immunodeficiency virus-1 antibodies: a technology whose time has come. Am. J. Med.102(Suppl. 4A), 9–14 (1997).
- Hodinka R, Nagashunmugam, T, Malamud, D. Detection of human immunodeficiency virus antibodies in oral fluids. Clin. Diagn. Lab. Immunol.5, 419–426 (1998).
- Sayer R, Law E, Connelly PJ, Breen KC. Association of a salivary acetylcholinesterase with Alzheimer’s disease and response to cholinesterase inhibitors. Clin. Biochem.37, 98–104 (2004).
- Streckfus C, Bigler L, Tucci M, Thigpen JT. A preliminary study of CA15–3, c-erbB-2, epidermal growth factor receptor, cathepsin-D, and p53 in saliva among women with breast carcinoma. Cancer Invest.18(2), 101–109 (2000).
- Streckfus C, Bigler L, Dellinger T, Dai X, Kingman A, Thigpen JT. The presence of soluble c-erbB-2 in saliva and serum among women with breast carcinoma: a preliminary study. Clin. Cancer Res.6(6), 2363–2370 (2000).
- Pepe MS, Etzioni R, Feng Z et al. Phases of biomarker development for early detection of cancer. J. Natl Cancer Inst.93(14), 1054–1061 (2001).
- Park NJ, Li Y, Yu T, Wong DT et al. Characterization of RNA in Saliva. Clin. Chem. (2006) (In Press).
Website
- The Oral Cancer Foundation www.oralcancerfoundation.org