Abstract
Triple-negative breast cancer (TNBC) is a complex heterogeneous disease characterized by the absence of three hallmark receptors: human epidermal growth factor receptor 2, estrogen receptor, and progesterone receptor. Compared to other breast cancer subtypes, TNBC is more aggressive, has a higher prevalence in African-Americans, and more frequently affects younger patients. Currently, TNBC lacks clinically accepted targets for tailored therapy, warranting the need for candidate biomarkers. BiomarkerBase, an online platform used to find biomarkers reported in clinical trials, was utilized to screen all potential biomarkers for TNBC and select only the ones registered in completed TNBC trials through clinicaltrials.gov. The selected candidate biomarkers were classified as surrogate, prognostic, predictive, or pharmacodynamic (PD) and organized by location in the blood, on the cell surface, in the cytoplasm, or in the nucleus. Blood biomarkers include vascular endothelial growth factor/vascular endothelial growth factor receptor and interleukin-8 (IL-8); cell surface biomarkers include EGFR, insulin-like growth factor binding protein, c-Kit, c-Met, and PD-L1; cytoplasm biomarkers include PIK3CA, pAKT/S6/p4E-BP1, PTEN, ALDH1, and the PIK3CA/AKT/mTOR-related metabolites; and nucleus biomarkers include BRCA1, the gluco-corticoid receptor, TP53, and Ki67. Candidate biomarkers were further organized into a “cellular protein network” that demonstrates potential connectivity. This review provides an inventory and reference point for promising biomarkers for breakthrough targeted therapies in TNBC.
Introduction
Breast cancer is the most common cancer in women, and the second most frequent cause of cancer-related deaths in women worldwide.Citation1 Approximately 20% of all breast cancers are referred to as triple-negative breast cancer (TNBC) due to a lack of three proteins: estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2).Citation2 TNBC tends to be more aggressive than other breast cancer subtypes and has a higher prevalence in African-Americans, more frequently affects younger patients (average age <50 years), and is associated with a greater risk of mortality.Citation1,Citation3 Currently, breast cancer treatment options and outcome are highly dependent on targeting ER, PR, or HER2. As a result, the American Society of Clinical Oncology (ASCO) does not currently recommend tailoring therapy for TNBC but instead recommends a general chemotherapy treatment based on the combination of an anthracycline with a taxane.Citation4
Although physicians have found that TNBC will initially respond to the combination of anthracycline and taxanes, treatment failure and disease recurrence continue to be clinically challenging.Citation5 Clinicians have attempted to tailor therapy for TNBC by monitoring the Food and Drug Administration (FDA)-approved biomarkers which can be classified as surrogate, prognostic, predictive, or pharmacodynamic (PD).Citation6 Surrogate biomarkers may predict an outcome by acting as a substitute for a clinical endpoint.Citation7 For example, evidence suggests that interleukin-8 (IL-8) blood levels are linked to breast cancer resistance protein (BCRP), a transient efflux transporterCitation8 that confers resistance to cytotoxic drugs;Citation9 therefore, IL-8 could be measured as a surrogate biomarker for BCRP levels in TNBC. Prognostic biomarkers suggest survival probability for cancer patients with and without drug therapy and would be used to ascertain whether patients require additional therapy.Citation6 For example, one study found that the presence of c-Kit, a protein expressed during cell replication, may determine the progression of TNBC.Citation10 Predictive biomarkers (also known as companion biomarkers) identify subpopulations of patients who are most likely to benefit from a specific drug therapy and form the basis for tailored-TNBC therapies. The HER2 protein is an example of a predictive biomarker that indicates a breast cancer that is more likely to have a favorable response to the drug trastuzumab (Herceptin; Genetech, Inc., South San Francisco, CA, USA).Citation4 Finally, PD biomarkers assess which molecular indicators are linked to a drug regimen, target effect, or tumor response, thereby providing rationale for new drug targets, drug combinations, or assay development. S6 and P4E-BP1 are potential PD biomarkers as their expression in in vitro TNBC experiments reflects activity in the TNBC chemotherapy-resistance pathway, PIK3CA/AKT/mTOR.Citation11
Although these four types of biomarkers provide a cornerstone of modern cancer therapeutics, they have not demonstrated significant clinical utility in the treatment of TNBC,Citation4 likely due to its highly complex biology and interpatient heterogeneity.Citation4 TNBC often exhibits resistance to standard combination chemotherapy through multiple interacting pathways with feedback and cross-talk loops.Citation6 These loops, in turn, may be altered by mutations in coding regions, regulatory elements, and noncoding sequences in the tumor DNA and gene expression,Citation12 which leads to downstream protein–protein, protein–gene, and other interactions which cannot be represented by single protein biomarker.
In the last decade, -omics technologies have been used to analyze the complex biology of TNBC by examining several forms of macromolecules (eg, DNA, RNA, proteins, and car bohydrates) that may shed light on TNBC signaling pathways.Citation13 These -omics technologies include proteomics, metabolomics, genomics, and epigenomics. Proteomic studies are used to measure the changes in tumor-associated proteins, which can differentiate between samples of normal tissue, benign, and malignant tumor or as any of the biomarkers described above. For example, the modification of specific proteins in patients with TNBC differentiates TNBC pathology and physiology from that of other forms of breast cancer.Citation14 In metabolomics, one analyzes how cells utilize small molecules (lipids, small peptides, and vitamins) to meet the energy requirements for sustaining life, and as building blocks for cellular division. For example, unlike healthy tissue, tumors perform aerobic glycolysis, thereby increasing lactate concentrations, which is known as the Warburg effect. In TNBC, this process appears to be interconnected with tumorigenic pathways PI3K/mTORCitation15 and EGFR,Citation16,Citation17 and metabolomic assays may be used to measure the response of TNBC to specific therapies that target these pathways.Citation13 Genomic studies of cancer cells examine genes specific to the tumor pathology which can include gene copy number, single nucleotide polymorphisms (SNPs), mutations, and loss of heterogeneity.Citation12 For example, deleterious BRCA1 mutations are found in high-risk TNBC populationCitation18–Citation20 and may increase tumor susceptibility to DNA-damaging and PARP inhibitor therapies.Citation21 Epigenomics is the examination of changes in cell phenotype that are the result of gene modification, such as DNA methylation, rather than changes in the DNA sequence itself.Citation22 For example, a significant proportion of TNBC may have BRCA1 promotor site hypermethylation;Citation18,Citation23,Citation24 although epigenetic silencing creates a similar protein profile to the loss-of-function BRCA1 mutation,Citation25 therapeutic efficacy may differ.Citation26
Aside from the complexity of TNBC, finding new and improved TNBC biomarkers is logistically challenging for several reasons. Centralized tumor specimen banks require proper sample collection, processing, and storage, which add financial burdenCitation27 and may deter candidate institutions from investing the necessary start-up capital. Following sample collection, data mining for novel biomarkers is time consuming and requires substantial input from data managers, bioinformaticians, and biostatisticians to correctly interpret the results.Citation6 Additionally, the biomarker discovery process is not always straightforward.Citation28 For example, because most cancer treatments use combination therapy rather than monotherapy, it can be difficult to connect the identified biomarker to a single drug or target.Citation6 Before a new biomarker can be implemented in the clinic, newly discovered TNBC biomarkers must be thoroughly examined and validated in order to potentially fill the gaps in our understanding of TNBC treatment and patient survival. In this work, biomarkers that have been studied in late-stage clinical trials were reviewed and were classified according to its biological location as blood (plasma or serum), cell surface, cytoplasm, or nucleus bio-markers. How recently published -omics studies may provide useful information on TNBC biomarkers is also discussed, and these markers are connected through an evidence-based molecular pathway landscape.
Methodology of data mining for biomarkers in TNBC
There are many preclinical study publications on TNBC bio-markers; a recent search in PubMed Central using the words “triple negative breast cancer and biomarker” returned over 2300 search results. In order to select only biomarkers with the most clinician-backed support, biomarkers associated with completed TNBC trials were chosen to be focused on by using BiomarkerBase, a biomarker knowledgebase™ developed by Amplion. BiomarkerBase uses a comprehensive list of synonyms to identify biomarkers registered in the records of clinical trials via the government website clinicaltrials.gov.
With BiomarkerBase, breast cancer biomarkers were first found through the search engine. Then, for each breast cancer biomarker, subsearches were conducted for clinical trials that explicitly used TNBC (or the full name, triple-negative breast cancer) in the title of the study. If the breast cancer biomarker was registered in at least one completed TNBC study, the biomarker was analyzed (with the exceptions of HER2, ER, and PR). Of note, most clinical trials surveyed for the work presented in this review completed Phase II or III. Current literature about the biomarkers was further examined using PubMed. Papers that studied one of the biomarkers as a general-disease biomarker, explored how -omics studies further characterized these biomarkers, and examined how the biomarker pathways may interact were sought.
Current advances in clinical biomarkers for TNBC patients
The following sections examine biomarkers found in the blood, on the cell surface, in the cytoplasm or nucleus in TNBC samples. Circulating blood biomarkers include vascular endothelial growth factor (VEGF), its receptor, VEGFR, and interleukin-8 (IL-8). The cell surface receptors include endothelial growth factor receptor (EGFR), insulin-like growth factor binding proteins (IGFBP), c-Kit, and PD-L1. All the plasma and cell surface biomarkers used in this review are associated with completed-TNBC clinical trials. Cytoplasm biomarkers include PIK3CA, pAKT/S6/p4E-BP1, PTEN, and PIK3CA/AKT/mTOR metabolites, in addition to ALDH1. PIK3CA, PTEN, ALDH1, and p4E-BP1 were registered in completed TNBC clinical trials, whereas pAKT/S6 biomarkers and the PIK3CA/AKT/mTOR metabolites were not. Nuclear biomarkers include BRCA1, the glucocorticoid receptor (GR), TP53, and Ki67 and were all studied in completed TNBC trials.
shows the system biology of TNBC. It summarizes all identified biomarkers through a “cellular protein network” that demonstrates the potential connectivity between the different subtypes of biomarkers. provides an overview of the examined surrogate, prognostic, predictive, and PD biomarkers in TNBC in addition to current literature found in PubMed that provides support for their potential use.
Figure 1 Biomarker pathways summarized through a “cellular protein network” that demonstrates potential connectivity.
Abbreviations: EGF, endothelial growth factor; ER, estrogen receptor; GC, glucocorticoid; GR, glucocorticoid receptor; HER2, human epidermal growth factor receptor 2; HGF, hepatocyte growth factor; HK2, hexokinase 2; IGFBP, insulin-like growth factor binding protein; IGF-R1, IGF receptor 1; IL-8, interleukin-8; mTOR, mammalian target of rapamycin; PDK1, phosphoinositide-dependent kinase 1; PIP2, phosphatidylinositol 4,5-biphosphate; PIP3, phosphatidylinositol 3,4,5-triphosphate; PKM2, pyruvate kinase M2; pPKM2, phosphorylated version of pyruvate kinase M2; PR, progesterone receptor; SPHK1, sphingosine kinase 1; VEGF, vascular endothelial growth factor; BCRP, breast cancer resistance protein.
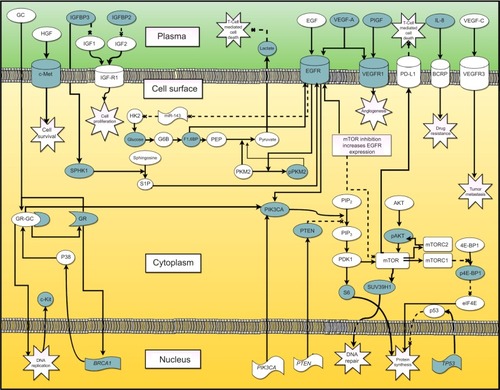
Table 1 Potential triple-negative breast cancer biomarkers
TNBC biomarkers in blood
Tumors or neighboring cells have been shown to release proteins that influence the survival of tumor cells,Citation3,Citation29 thereby leading to a worse outcome in TNBC patients. Only few biomarkers were reported in the serum or plasma of TNBC patients in late-stage clinical trial, and so far, none has been approved by the FDA, yet. They include VEGF, VEGFR, and IL-8.
VEGF/VEGFR
VEGF is typically found in blood as a disulfide-linked homodimer. Created from a family of six proteins, these VEGF isoforms include VEGF-A, VEGF-B, VEGF-C, VEGF-D, VEGF-E, and placental growth factor (PlGF). Glycosylated VEGF binds the VEGFR, a family of receptor tyrosine kinases (RTKs) including VEGFR1, VEGFR2, and VEGFR3, which upon stimulation lead to an increase of angiogenesis and the permeability of nearby vessels and lymphatics;Citation3,Citation30,Citation31 the end result of which is improved oxygen/nutrient transport, as well as increased tumor metastasis.Citation32,Citation33
Roberti et alCitation34 demonstrated the potential use for VEGF as a prognostic biomarker in TNBC. Briefly, animal models were implanted with either metastatic or nonmetastatic TNBC cells. The metastatic TNBC cells tended to express higher levels of PlGF and VEGF-A than the nonmetastatic counterparts, suggesting that overexpression of these markers correlated with metastatic potential for TNBC. Later, in a clinical study conducted by Bahhnassy et al,Citation35 VEGF-A expression was examined before and after standard chemotherapy. In agreement with Roberti’s study, the expression of VEGF-A correlated with a poorer overall survival and a less-favorable response to chemotherapy.
Overwhelming preclinical evidence predicted a positive response to bevacizumab (anti-VEGFA therapy) in TNBC patients, and thus, the potential for VEGF as a predictive biomarker for VEGF-tailored treatment;Citation36,Citation37 unfortunately, the clinical results were not positive. Bear et alCitation38 examined breast cancer response to the combination of bevacizumab and standard chemotherapy. The addition of bevacizumab significantly increased the overall rate of pathological complete response (pCR), but after subset analysis, bevacizumab was found to be slightly more effective in the HER2-positive group than the TNBC group.
Shortly after Bear et al released their results, Chen et alCitation21 released a meta-analysis examining eleven studies focused on standard neoadjuvant chemotherapy with- and without bevacizumab in TNBC patients. The meta-analysis revealed that, overall, TNBC treated with bevacizumab resulted in an improvement in pCR.Citation21 Interestingly, Chen et al found no clear homogeneity between the studies, suggesting that the current studies do not provide enough patient details including VEGF-A expression to determine which TNBC subgroups most benefit from VEGF therapy.
VEGFR levels may be indicative of successful anti-VEGF-A therapy in TNBC. Recently, Tolaney et alCitation39 conducted a Phase II study to examine the effect of bevacizumab on TNBC patients and found that TNBC with high VEGFR1 had the greatest response to anti-VEGF-A therapy. The therapy appeared to prune vessels high in VEGFR1 while normalizing the other vessels, suggesting that high baseline VEGFR1 microvascular density may be required for successful neoadjuvant anti-VEGF-A therapy in TNBC. TNBC may be dependent on both VEGF and VEGFR analysis for effective treatment, and VEGF and VEGFR together as predictive biomarkers may demonstrate improved clinical utility for tailored TNBC therapy.
IL-8
IL-8 (or CXCL8) is a major CXC motif cytokine encoded by IL-8 on chromosome 4q13-q21. IL-8 is produced by TNBC tumors in response to hypoxic conditions and is thought to recruit mesenchymal stem cells (MSCs) to the TNBC location.Citation9 MSCs normally reside in the bone marrow and adipose tissue, but when recruited by TNBC, they home to the breast cancer tumor location and create a microenvironment around the tumor hypothesized to increase the stem-cell-like characteristics of the TNBC,Citation29 thereby increasing TNBC multidrug resistance (MDR) and metastatic risk.Citation34
TNBC often becomes resistant to doxorubicin, a standard anthracycline used in TNBC treatment.Citation40 TNBC resistance to doxorubicin may be due to the ability of IL-8 to upregulate the BCRP found on the surface of TNBC cells. BCRP is a 72 kDa transmembrane protein responsible for removing doxorubicin from a tumor cell.Citation8,Citation41–Citation43 In vitro data suggest that baseline levels of BCRP expression on the cell surface is very high in TNBC, and the expression can be further upregulated in response to drug therapy.Citation8 Importantly, BCRP upregulation is transient, lasting only for a few hours after exposing the tumor to doxorubicinCitation8 while IL-8 expression can last for several days.Citation44 The potential usefulness of IL-8 as a biomarker was demonstrated by an in vitro study by Chen et al,Citation9 which demonstrated that IL-8 increased BCRP expression without affecting the expression of other hallmark efflux transporters correlating with increased TNBC resistance to doxorubicin. IL-8 may serve to protect the TNBC from doxorubicin-induced killing by increasing BCRP levels in TNBC and may function as a surrogate biomarker for BCRP expression in TNBC.
TNBC biomarkers on the cell surface membrane
Various membrane-bound receptors have the potential function as biomarkers for TNBC as they were shown to increase anti-apoptotic signals in TNBC cell lines; thus, their blockade would be expected to increase tumor death either alone or as part of combination therapy. These biomarkers include EGFR, IGFBP, C-kit, and PD-L1.Citation46–Citation48 In this section, each of these membrane-bound candidate biomarkers are described along with the clinical trials in which they were investigated.
EGFR
The EGFR family consists of four similarly structured RTKs that have important roles in tumor proliferation: EGFR (HER1), HER2, HER3, and HER4. After EGF or a related ligand such as tumor growth factor α binds to the EGFR, the EGFR dimerizes resulting in activation of the EGFR intracellular tyrosine kinase (TK) domain, which in turn recruits additional linker molecules and intracellular TKs. This induces an intracellular signaling cascade that promotes cell proliferation, angiogenesis, metastatic spread, and apoptotic inhibition. Of the four EGFR receptor family members, TNBC most frequently expressed EGFR/HER1.Citation46
Significant evidence shows that EGFR overexpression in TNBC makes TNBC more difficult to treatCitation49 and significantly lowers the 10-year survival rate in breast cancer patients.Citation50 Although based on preclinical data, anti-EGFR therapy would seem to have an application in TNBC, clinical results have so far been disappointing in trials,Citation51 and as such, the role of EGFR in TNBC therapy has been a hot topic for debate for the past two decades.Citation49 The Asian population might be a special case; however, the studies making these claims do not have an agreed upon mechanism of action. In a genomic analysis of 110 Japanese TNBC patients, 30% of the TNBC had EGFR overexpression, with 21% having an EGFR copy number increase. Interestingly, there was no correlation between EGFR expression and EGFR copy number.Citation52 A later study by Teng et alCitation53 examined 70 Asian TNBC tumors (predominantly Chinese) and found 12% of the tumors to carry an EGFR mutation. At first glance, the EGFR mutations found in Teng et al’sCitation53 study appeared to be unique to Asian TNBC as they were not found in any European TNBC study conducted during the same year.Citation54 However, like the EGFR copy number changes in the previous study, EGFR mutations did not appear to account for the increased EGFR expression in Asian patients. The biology behind EGFR expression may not be completely explained by genomics. Nevertheless, the EGFR may represent a useful target in Asian patients as EGFR mutations, but not EGFR levels, signal responsiveness to EGFR-targeted therapy in non-small cell lung cancer.
A recent landmark preclinical study by Lim et alCitation17 utilized metabolomics and proteomics to shed light on an EGFR unique mechanism of action in TNBC. Unlike the metabolism in healthy tissue, many tumors metabolize glucose via the aerobic glycolysis pathway. The study by Lim et alCitation17 examined two vital isozymes that regulate aerobic glycolysis in tumor proliferation: hexokinase 2 (HK2) and pyruvate kinase M2 (PKM2). HK2 catalyzes glucose phosphorylation, the first step in the aerobic glycolysis pathway. EGFR “removes the breaks” from HK2 expression by downregulating miR-148, a microRNA that normally silences HR2 expression. Increased HK2 expression increases glucose utilization. PKM2, an embryonic isozyme, phosphorylates phosphoenolpyruvate into pyruvate, thereby accelerating the last step in aerobic glycolysis. EGFR activity leads to PKM2 phosphorylation at the Tyr148 residue, forming a slightly less active phosphorylated version (pPKM2)Citation17 and thereby slowing down the last step in the aerobic glycolytic pathway.
Increased HK2 activity combined with decreased pyruvate production via pPKM2 creates a “glycolytic jam” where intermediate aerobic glycolytic metabolites accumulate and have the potential to drive the production of nonessential amino acids needed for cellular proliferation. Interestingly, the metabolite fructose 1,6-bisphosphate (F1,6BP) was found to independently alter the phosphorylation of EGFR, thereby creating a positive feedback loop. Because the aerobic glycolytic pathway is active, there is an increase in extracellular lactate production. Lim et alCitation17 found that pPKM2 was positively correlated with increased EGFR expression and increased Ki67 levels, a generalized biomarker for cell proliferation. Because these characteristics were found only in the TNBC lines examined,Citation17 extracellular lactate, F1,6BP, and pPKM2 may have roles as predictive biomarkers in EGFR expressing TNBC.
IGFBP
IGFBPs are a family of six receptors (IGFBP1–IGFBP6) that regulate tumorigenesis through binding to insulin-like growth factors (IGF) both increasing IGF half-life and sequestering IGF.Citation55 IGF, in turn, binds and stimulates IGF receptor 1 (IGF-R1), which leads to proliferative and antiapoptotic effects through activating the PIK3/AKT pathway.Citation47,Citation56 IGFs are secreted by adipocytes as well as cancer cells and have been proposed to increase the risk of breast cancer metastasis.Citation57 African-Americans have a higher prevalence of obesity and TNBC risk compared to Caucasians,Citation1,Citation58 which may be due to a contribution of the IGFBP/IGF pathway in TNBC.
IGFBP2 is a fetal growth factor overexpressed in neoplastic cells,Citation56 especially in HER2-negative breast cancer.Citation59 Mechanistic studies examining the role of IGFBP2 in TNBC are limited, but current evidence suggests that IGFBP2 binds both IGF1 and IGF2 and increases the opportunity for IGFs to bind to IGF-1R.Citation47
There is conflicting evidence on IGFBP2 as a TNBC marker. Preclinical evidence suggests that IGFBP2 may function as a potential prognostic biomarker.Citation34 In addition, IGFBP2 was found to be a predictor of recurrence-free survival (RFS) when measured along with four other proteins in TNBC patients receiving post-neoadjuvant chemotherapy.Citation60 However, a report by Hernandez et alCitation61 in 2015 disagreed with the findings. Hernandez et al examined the relationship between IGF and IGFBP expression with survival rates in Asian, Pacific Islander, and Caucasian patients and found that increased IGFBP2 expression correlated to a decreased breast cancer survival rate, and the increased IGFBP2 expression varied between racial/ethnic groups. Additionally, this study showed that TNBC was associated with decreased IGF1 and IGFBP2 expression.Citation61 The African-Americans, the demographic with the highest TNBC risk, were not tested. IGFBP2 may have utility in combination with other biomarkers as a prognostic biomarker in ethnic populations; however, more evidence for an IGFBP2 correlation with outcomes in the TNBC in the African-American population is needed.
IGFBP3 has a similar mechanism of action to IGFBP2 by binding to IGF-1 and IGF-2 and increasing its half-life. IGF, in turn, bind to IGF-1R, leading to increased tumor survival. Studies disagree whether IGF-1 and IGF-1R are correlated with the presence of TNBC.Citation35,Citation61 Because of the uncertainty of this mechanism, IGFBP3/IGF-1 pathway may be a part of a larger cellular process as IGFBP3 appears to be independently associated with aggressive TNBC.Citation55 Increased IGFBP3 expression appears to increase EGFR expression through sphingosine kinase 1 (SPHK1) activity in TNBC,Citation62,Citation63 and as mentioned in the EGFR section, the EGFR can lead to increased cell proliferation, angiogenesis, and tumor metastasis. As such, IGFBP expression may function as a prognostic biomarker, and SPHK1 may function as a PD biomarker.
c-Kit
c-Kit (also called CD117 and stem cell factor receptor) is an RTK encoded by the 21 exon proto-oncogene, c-Kit, located on chromosome 4q12. c-Kit is found on the surface of hematopoietic stem cells, and after binding to its substrate cytokine, it increases cell survival proliferation and chemotaxis.Citation48 The use of c-Kit as a TNBC biomarker has been suggested, but is unclear. c-Kit may be expressed in ~25%–45% of TNBC,Citation64,Citation65 but studies disagree as to whether c-Kit levels predict the overall survival of TNBC patients.Citation10,Citation66,Citation67 A Phase II trial examined sunitinib (a targeted agent that inhibits c-Kit as well as multiple other kinases) as monotherapy in patients with advanced TNBC as compared to standard chemotherapeutic regimen and found that sunitinib was not an effective treatment.Citation68 Because of the controversial evidence of c-Kit in TNBC, more studies are needed in order to determine its potential usefulness as a TNBC biomarker.
c-Met
c-Met (also called hepatocyte growth factor receptor [HGFR]) is an RTK that, after binding to its substrate hepatocyte growth factor (HGF), increases cell survival. c-Met is encoded by the proto-oncogene c-Met on chromosome 7q21-31, and, while c-Met mutation, amplification, or c-Met overexpression leads to increased proliferation, motility, and invasion of cancerous tissue,Citation69 increased c-Met copy number appears to be the most common origin of c-Met aberrations in TNBC.Citation70,Citation71
Contrary to preclinical evidence, recent clinical data suggest that c-Met inhibition may not demonstrate utility as a TNBC treatment option.Citation39,Citation72,Citation73 Dieras et alCitation73 examined metastatic TNBC treated with the addition a c-Met inhibitor, onartuzumab, alongside bevacizumab or paclitaxel. The addition of the c-Met inhibitor did not improve patients’ progression-free survival nor their overall survival. Similarly, Tolaney et alCitation39 examined metastatic TNBC, except the subjects that were treated solely with the oral c-Met inhibitor, tivantinib. Toxicity from the c-Met inhibitor was minimal, but the c-Met inhibitor monotherapy treatment was not efficacious. Although treating TNBC inhibitor does not appear to be as effective as once thought, a meta-analysis conducted by Yan et alCitation74 demonstrated that overexpression of c-Met increases the risk of RFS in TNBC. c-Met may, therefore, function as a prognostic biomarker in TNBC patients.
PD-L1
Programmed cell death 1 ligand 1 (PD-L1, B7-H1, CD274) is a transmembrane protein encoded by CD274 that functions as a key checkpoint regulator in the immune response.Citation75,Citation76 PD-L1 is typically found in B cells, natural killer cells, and vascular endothelial cells and binds the programmed cell death protein 1 (PD-1) found on activated cytotoxic T-cells. PD-L1/PD-1 binding prevents the release of IL-2, T-cell activation, and proliferation, thereby serving as an important regulatory checkpoint preventing excessive adaptive immune responses.Citation75
PD-L1 expression on tumor cells appears to be higher in TNBC than non-TNBCCitation75,Citation77 and is estimated to occur in ~20% of TNBC.Citation78 A recent clinical trialCitation79 used pembrolizumab, a high-affinity anti-PD-L1 antibody in metastatic, PD-L1 expressing TNBC patients. The overall survival in patients on pembrolizumab monotherapy was 18.5%, demonstrating the potential role of PD-L1 as a predictive TNBC biomarker in tailoring immune checkpoint therapy.
The exact pathway by which PD-L1 is upregulated in TNBC is not fully understood. A preclinical study by Mittendorf et alCitation78 suggests that PD-L1 expression may be associated with PTEN loss. Mittendorf et alCitation78 compared PD-L1 expression in PTEN-knockdown TNBC lines to non-PTEN knockdown lines. As mentioned in the “PIK3CA” section, two downstream targets of PTEN are AKT and mTOR.Citation80 When non-PTEN knockdown lines were treated with either AKT inhibitor or mTOR inhibitor, they exhibited inhibited PD-L1 expression while the PTEN knockdown demonstrated increased PD-L1.Citation78 As a result, PD-L1 expression may be linked to the activation state of the PIK3CA pathway.
Another potential mechanism by which PD-L1 is upregulated in TNBC is through interferon γ (IFNγ), an inflammatory mediator.Citation75,Citation81 Soliman et alCitation75 noted two proteins that positively and negatively regulate IFNγ expression, STAT1 and interferon regulatory factor 2 binding 2 protein 2 (IRF2BP2), respectively. The study found that breast cancer cell lines with the highest PD-L1 expression tended to have higher levels of STAT1 and lower levels of IRF2BP2 expression. Although not yet tested clinically, a high STAT1 to low IRFBP2 ratio may identify TNBC that have higher PD-L1 upregulation potential and may function as a surrogate bio-marker for response to checkpoint inhibitors.
TNBC biomarkers in the cell cytoplasm
Majority of the biomarkers in the cytoplasm of TNBC cells have been shown to confer TNBC resistance to drug therapy in interventional studies.Citation82–Citation84 These biomarkers include proteins in the PIKCA/AKT/mTOR pathway such as PIK3CA, PTEN, pAKT/pS6/p4E-BP1, and associated metabolites in addition to ALDH1. In this section, these proteins are all detailed along with the clinical trials in which they were investigated.
PIK3CA/AKT/mTOR
The PIK3CA/AKT/mTOR pathway has gained popularity as a potential route for TNBC resistance to chemotherapy.Citation82,Citation83PIK3CA, at the top of the pathway, is located in a span of DNA on chromosome 3q that encodes for the p110α catalytic subunit of the phosphoinositide 3-kinase 3A (PIK3CA) among other mediators and is amplified in a significant fraction of TNBC. PIK3A is an upstream catalytic enzyme that, when active, leads to cell growth and proliferation and in particular inhibition of cell death. PIK3CA activating mutations, as well as general dysregulation of the PIK3CA/AKT/mTOR pathway, are associated with TNBC.Citation85,Citation86 PIK3CA phosphorylates phosphatidylinositol 4,5-biphosphate (PIP2) to form 3,4,5-triphosphate (PIP3), which activated multiple proteins containing PH and other domains that are recruiting to PIP3, thereby leading to the activation of a downstream signaling cascade that mediates the effects of PIK3CA including cell proliferation. PIP3 recruits phosphoinositide-dependent kinase 1 (PDK1) and AKT to the cell membrane among other PH domain proteins, and when PDK1 and AKT are in close proximity, PDK1 phosphorylates AKT (pAKT) thereby increasing AKT activity.Citation87
pAKT/pS6/p4E-BP1
pAKT activates mammalian target of rapamycin (mTOR), a key serine/threonine kinase that is vital for TNBC cell survivalCitation46 and proliferation.Citation88 mTOR regulates three important proteins in the PIK3CA/AKT/mTOR pathway: S6, eukaryotic translation initiation factor 4e binding protein (4E-BP1), and SUV39H1. S6 is a 40S ribosomal protein that regulates translation and is frequently used in determining the downstream activity of an important therapeutic target, the mTOR complex (mTORC1).Citation11
Activated mTOR leads to downstream 4E-BP1 phosphorylation (p4E-BP1), which stimulates cap-independent translation.Citation89 Cap-independent translation is a potential mechanism by which large breast tumors stimulate angiogenesis under hypoxic conditions.Citation90 Studies on large breast tumors found that 4E-BP1 expression was positively associated with cell survival, demonstrating p4E-BP1’s role as a potential prognostic biomarker.Citation91–Citation93 p4E-BP1 can also be used as an indication of the activity of the therapeutic target mTOR complex 1 (mTORC1). When mTORC1 is inhibited, mTORC2 activity increases and vice versa. pS6 and p4E-BP1 have demonstrated usefulness as dual PD biomarkers for determining mTOR pathway activity.Citation11
Finding TNBC-specific biomarkers through the mTOR pathway has been challenging, as mTOR expression and activity seem to be similar in TNBC and non-TNBC.Citation94 Nascent chromatin capture is a relatively new biochemical process that recently uncovered SUV39H1, a methyltransferase, as a potential protein biomarker.Citation95 SUV39H1 modulates DNA expression through histone modification, an epigenetic route of action, which seems to play a major role in homologous recombination (HR), a key DNA damage repair pathway and a determinant of sensitivity to PARP inhibitors and platinum-based chemotherapy.Citation96 SUV39H1 may bridge mTOR activity with BRCA1 activity. As mentioned in the “BRCA1” section, many TNBCs have aberrant BRCA1 and BRCA2 function with these proteins playing a critical role in HR in double-strand break repair.Citation97 Inhibiting mTOR leads to SUV39H1 suppression, and the decreased SUV39H1 has been correlated to decreased DNA repair in BRCA1-mutated TNBC samples, independent of pS6 and p4E-BPT.Citation11 SUV39H1 may prove to be useful proteomic/epigenomic PD biomarker in tailored TNBC therapy targeting DNA repair.
PTEN
PTEN also represents the key negative regulator of PIK3CA/AKT/mTOR signaling. PTEN is a nine-exon tumor suppressor gene found on chromosome 10q23 that encodes PTEN, a dual acting phosphatase.Citation80 PTEN dephosphorylates the 3-phosphoinositide products of PI3Ks, acting as a break for the PI3K/AKT/mTOR pathway, and thus inhibits TNBC metastasis and replication.Citation80PTEN is considered a high-penetrance breast cancer predisposing gene because PTEN mutations found in Cowden’s syndrome are associated with and increased lifetime breast cancer incidence.Citation98 PTEN SNPs have also been associated with TNBC incidence.Citation99 PTEN loss, which can occur through multiple mechanisms, has been associated with breast cancer tumor size, grade, reoccurrence, drug resistance, and worse prognosis.Citation100
A proteomic study on 76 breast cancer biomarkers examined proteins that could predict RFS in TNBC patients. The study created a risk score (RS) module composed of six proteins. Three were proteins from the PIK3/AKT/mTOR pathway including AKT, S6, and PTEN. The other three proteins were IGFBP, stathmin, a regulator of cell division, and LKB1, a kinase-activating kinase.Citation60 As mentioned previously in the “PD-L1” section, PTEN loss may be associated with greater PD-L1 expression,Citation78 which may help to explain the role of PTEN in TNBC outcomes.Citation99,Citation100
Metabolites
Metabolomics may play an important role in examining the functional consequences of PIK3CA/AKT/mTOR pathway activity. The PIK3CA/AKT/mTOR pathway is a key regulator of glucose uptake. As discussed in the “EGFR” section, tumors frequently have high levels of aerobic glycolysis unlike normal tissues, thereby increasing extracellular lactate concentrations. Moestue et alCitation15 found that inhibition of PI3K/mTOR in TNBC decreased lactate production and increased glucose levels. In addition, Chen et alCitation16 found that inhibiting mTOR also leads to EGFR upregulation in TNBC, which has shown to increase lactate production in a recent study by Lin et alCitation17 albeit in the absence of PI3K pathway inhibition. This evidence suggests that lactate/glucose levels may serve as an important metabolomic PD biomarker in monitoring PI3KCA/AKT/mTOR activity, as well as its relationship to EGFR signaling in TNBC-directed therapy.
Choline metabolism may also be selectively important for TNBC. Moestue et alCitation15,Citation101 found that PI3K/mTOR inhibition leads to altered choline metabolism in basal-like breast cancer. Glycerophosphocholine (GPC) to phosphocholine (PChO) conversion was significantly higher in aggressive basal-like breast cancer.Citation101 Because of the high overlap between TNBC and basal-like breast cancer,Citation50 GPC and PCh may function as prognostic biomarkers in TNBC.
ALDH1
ALDH1 catalyzes the oxidation of endogenous and exogenous aldehydes into inactive carboxylic acid species.Citation102 ALDH1 is a cytoplasmic stem cell-related marker found in a number of breast cancers and is associated with tumor initiating cells.Citation103 ALDH1 expression is significantly correlated with tumor grade metastasisCitation104 and may be associated with increased resistance to taxane- and epirubicin-based chemotherapy.Citation84
ALDH1 may have promise as a TNBC-specific marker. Ohi et alCitation105 examined 107 TNBC tumors that express EGFR and cytokeratin 5/6 (this phenotype was described as basal-cell TNBC by Kashiwagi et alCitation10). The study found that relapse-free survival was lower in ALDH1-positive tumors, suggesting its potential role as a prognostic biomarker in TNBC. Another study examined 147 invasive breast tumors from African patients from Ghana and found an association between the prevalence of ALDH1 expression in TNBC and androgen receptor (AR) expression.Citation106 AR expressing TNBC cell lines are more sensitive to AR antagonists leading to trials exploring the role of AR antagonists in TNBC. ALDH1 is thus a potential predictive biomarker for AR-targeted therapy in TNBCCitation107 as well as a likely prognostic marker.
TNBC biomarkers in the cell nucleus
A number of nuclear biomarkers including ER and PR have a well-established role as prognostic and predicted biomarkers in breast cancer. The role of nuclear proteins as biomarkers in TNBC is less well developed. A number of nuclear biomarkers including BRCA1/2, GR, and TP53 have been clinically validated as risk factors for cancer development,Citation108 tumor survivability,Citation85 and tumor proliferation.Citation109 In this section, each of these genes and the clinical trials in which they served as biomarkers of disease were described.
BRCA1 and BRCA2
BRCA1 and BRCA2 are protein-expressing spans of DNA found on chromosome 17q21 and 13:12.3, respectively. As noted earlier, these proteins are critical components of the HR DNA repair pathway. Mutations in BRCA1 and BRCA2 have been linked to increased lifetime breast cancer incidence, independent of other breast cancer-related genes.Citation110BRCA1 and BRCA2 are considered a high-penetrance breast cancer-predisposing gene because of the strong correlation between BRCA1 aberrant genetic changes (either genomic or epigenomic) and TNBC risk.Citation108
Evidence suggests that BRCA1-mutated breast cancer has significant overlap with TNBC. BRCA1-related breast cancers share pathological features with TNBC (a phenotype called “BRCAness”) including low or changed expression of ER/PR/HER2, EGFR expression, TP53 mutation, extreme genomic instability from HR deficiency, and a high mitotic index.Citation111,Citation112BRCA1-associated breast cancer also shares two unique metastasis characteristics with TNBC. Although most breast cancer metastasis risks typically correlate with increasing tumor size, there is no apparent association between BRCA1-related breast cancer metastasis risk and tumor size, which is also seen in TNBC. In addition, most breast cancer metastasis risk remains constant over time, but in BRCA1-negative breast cancer and TNBC, metastasis risk seems to increase significantly after 3 years, then decline rapidly thereafter.Citation113,Citation114
The exact mechanism by which BRCA1 and BRCA2 loss contributes to breast cancer predisposition is still unknown and in particular why individuals with germline mutations are prone to specific subset of tumors such as breast and ovarian cancer. However, the major role is probably due to the defects in HR and gene transcription, and thereby a decreased efficiency in repairing double-strand DNA breaks.Citation108BRCA1 and BRCA2 loss leads to non-HR repair,Citation111 thereby increasing genome instability and tumor mutation.Citation97 Because BRCA1 and BRCA2 seem to be heavily involved in DNA repair, aberrations in BRCA1 and BRCA2 appear to sensitize TNBC to DNA-damaging platinum agents and PARP inhibitors. A meta-analysis from Chen et alCitation21 examined the risk of remission rate in TNBC using standard neoadjuvant therapy with- or -without carboplatin. The results suggest that carboplatin improves pCR over than other agents used in TNBC treatment.
BRCA1- and BRCA2-related TNBC is categorized into two groups, familial or sporadic, both of which exhibit the “BRCAness” phenotype. The term “familial” makes the assumption that the cancer occurs due to a germline predisposition aberration.Citation115 Clinically, TNBC is classified as familial if the TNBC patient meets the following criteria: the patient has at least three breast and/or ovarian cancer cases in the family, two TNBC cases in close relatives with at least one diagnosed before 50 years old, or two breast cancer cases in the family before 40 years old, Ashekenazi Jewish ancestry, or has ovarian cancer in addition to their TNBC.Citation108 Germline BRCA1 and BRCA2 mutations represent the majority of BRCA1- and BRCA2-related TNBC.Citation116–Citation118 Majority of familial TNBC may be caused by deleterious germline BRCA1 and BRCA2 mutationsCitation18 as seen in young TNBC patients (<40 years old) with aggressive TNBCCitation19 and Ashkenazi Jewish TNBC patients.Citation20 However, current data suggest that the potential likelihood of an individual with a relatively early onset TNBC having a germline aberration in BRCA1 and BRCA2 is sufficiently high that genetic counseling and genomic testingare warranted.
Only a small proportion of TNBC can be explained by germline aberrations or family history.Citation115 Approximately 7% of breast cancers have somatic mutations in BRCA1 and BRCA2.Citation99,Citation119 Epigenetic silencing of BRCA1 but not BRCA2 may contribute to sporadic TNBC. Epigenetic BRCA1 silencing occurs through hypermethylation of CpG islands in the BRCA1 promotor sequence.Citation23 Hsu et alCitation24 examined 139 early stage Taiwanese breast cancer patient tissues and demonstrated an association between hypermethylation of the BRCA1 promoter and TNBC. This idea is reinforced with evidence from a recent study by Zhu et alCitation18 that found 50% of sporadic TNBC cases also demonstrated promotor site hypermethylation.Citation120 However, there is a conflicting evidence regarding the significance of CpG island methylation in TNBC.Citation121
Epigenetics may be an important factor in tailoring treatment options. One study suggests that a standard anthracycline plus taxane regimen may be more effective in sporadic TNBC with BRCA1 promotor site hypermethylation than those without hypermethylation.Citation26 In contrast, another study suggests that the taxane regimens may not be effective in BRCA1 promotor site hypermethylated TNBC but that platinum agents are more effective in treating this subgroup.Citation122 Because epigenetic silencing BCRA1 silencing creates a similar BRCA1 expression profile as BRCA1-mutant breast cancer,Citation25 epigenetic tests are needed to verify BRCA1 activity and tailor therapy for TNBC. Although mutations in BRCA1 and BRCA2 are clearly associated with prognosis and likely prediction of responsiveness to platinum and PARP inhibitor therapy, it remains unclear whether BRCA1 silencing plays a similar role potentially due to silencing being reversible under therapeutic stress.
GR
GR is encoded by a nine-exon span of DNA located on 5p31q.Citation123 The GR ligand, glucocorticoid (GC), is a protein-bound plasma hormone released from the adrenal cortex during times of stress. Ligand-activated GR translocates to the nucleus where it dimerizes and increases the transcription of GC-inducible genes,Citation124 which leads to antiapoptotic activity and resistance to chemotherapy in TNBC.Citation125,Citation126
The mechanism by which GR acts has been debated. Preclinical evidence suggests that GR antiproliferative effects are mediated by BRCA1, where BRCA1 activity leads to downstream phosphorylation of the MAPK p38, which, in turn, phosphorylates GR to GR-active and GC independent form (P-Ser211).Citation124 However, some studies suggest that GR long-term activity decreases BRCA1 expression while free GR increase BRCA1 expression.Citation127,Citation128 More evidence is required for the specifics of this mechanism, but GR and P-Ser211 may be useful proteomic PD biomarkers in TNBC therapy.
TP53
TP53 is coded by a gene located on the chromosome 17p13, encoding the tumor suppression protein p53. Cellular stress induces p53 expression, which induces cell cycle arrest, apoptosis, DNA repair, and cell-metabolism changes.Citation85TP53 that is mutated in the germline of Li Fraumeni families is a high-penetrance breast cancer disposing gene with mutations in this segment being associated with a high risk of breast cancer and in particular very early onset breast cancer.Citation129 Genomic TP53 alterations are prevalent in TNBC tumorsCitation99,Citation122 with up to 85% of basal breast cancers having TP53 mutations.Citation45,Citation130
There has been a debate over the characteristics of TP53 mutations in TNBC. A study by Kim et alCitation45 found that neither frameshift, nonsense, nor splicing TP53 mutations were associated with breast-specific survival in TNBC. Rather, the study reported that missense DNA binding motif mutations and non-DNA-binding motif mutations were associated with a higher rate for disease relapse. Other studies however propose that loss of TP53 function regardless of the underlying genomic event is associated with worse clinical outcome. Indeed, these studies found no significance in the type of TP53 mutation.Citation122,Citation130 Foedermayr et alCitation122 found that most of the TP53 mutations in TNBC were localized in the DNA-binding domain. A number of recent studies suggest that decreased TP53 function is associated with worse overall survival in TNBC patientsCitation130 and increased metastatic risk.Citation85 A recent study by Powell et alCitation85 suggests that TP53 interacts with the BTG2 promoter, which functions to enhance tumor proliferation. A BTG2 role as a PD biomarker is unclear; however, TP53 mutation may be important as a prognostic biomarker in aggressive TNBC.Citation45 The discrepancies between studies on the role of p53 as a prognostic marker in TNBC may arise from suggestions that the function of the p53 pathway is abnormal in the majority of basal breast cancer due to aberrations in TP53 as well as other critical members of the pathway. If tumors with TP53 mutations represent only a subset of tumors where the p53 pathway is aberrant, TP53 mutation alone would not be expected to be a powerful biomarker
Conclusion
Breast cancer is a complex, heterogeneous disease currently categorized by the expression of three predominate receptors, namely ER, PR, and/or HER2, as prognostic and predictive biomarkers. The current recommendations by the ASCO suggest to treat postmenopausal women with ER-expressing metastatic tumors with an aromatase inhibitor such as anastrozole, letrozole, or exemestane, and premenopausal patients with ER-expressing metastatic tumors with a selective ER modulator such as tamoxifen, raloxifene, or toremifene. Likewise, the ASCO suggests to treat HER2-expressing metastatic tumors with trastuzumab, an anti-HER2 monoclonal antibody.Citation4 To date, there are no clinically validated biomarkers for TNBC, which has hindered the developmentCitation4 of tailored therapy for both chemosensitive and refractory TNBC.Citation5
TNBC biology is a complex interplay of protein–protein, protein–DNA, or other component–component interactions. Indeed, TNBC is the most heterogenous of all types of breast cancer being composed of multiple different subtypes.Citation107 Omics technologies provide cutting-edge tools for an in-depth understanding of the molecular landscape of TNBC, where data are extracted from a clinical source, a network between these pathways is inferred, and -omic biomarkers developed through the process of top-down modeling.Citation2,Citation131 However, validation of these biomarkers must occur, and stringent-clinical criteria met before we can rely on their routine use in the clinical setting. The list provided is an inventory and reference point for promising biomarkers for breakthrough, targeted therapies in TNBC.
Disclosure
The authors report no conflicts of interest in this work.
Acknowledgments
We want to thank Dr Gordon Mills at MD Anderson Cancer Center for reviewing this manuscript and sharing his knowledge of biomarkers and TNBC.
References
- SiegelRMaJZouZJemalACancer statistics, 2014CA Cancer J Clin201464192924399786
- PodoFBuydensLMDeganiHFEMME Consortium. Triple-negative breast cancer: present challenges and new perspectivesMol Oncol20104320922920537966
- BenderRJMac GabhannFExpression of VEGF and semaphorin genes define subgroups of triple negative breast cancerPLoS One201385e6178823667446
- Van PoznakCSomerfieldMRBastRCUse of biomarkers to guide decisions on systemic therapy for women with metastatic breast cancer: American society of clinical oncology clinical practice guidelineJ Oncol Pract2015332426952704
- PerouCMMolecular stratification of triple-negative breast cancersOncologist201116Suppl 16170
- de GramontAWatsonSEllisLMRodónJTaberneroJde GramontAHamiltonSRPragmatic issues in biomarker evaluation for targeted therapies in cancerNat Rev Clin Oncol201512419721225421275
- LipsEHMichautM3HoogstraatMCenter for Personalized Cancer Treatment. Next generation sequencing of triple negative breast cancer to find predictors for chemotherapy responseBreast Cancer Res201517113426433948
- DufourRDaumarPMounetouEBCRP and P-gp relay overexpression in triple negative basal-like breast cancer cell line: a prospective role in resistance to OlaparibSci Rep201551267026234720
- ChenDRLuDYLinHYYehWLMesenchymal stem cell-induced doxorubicin resistance in triple negative breast cancerBiomed Res Int2014201453216125140317
- KashiwagiSYashiroMTakashimaTc-Kit expression as a prognostic molecular marker in patients with basal-like breast cancerBr J Surg2013100449049623319435
- MoWLiuQ1LinCCmTOR inhibitors suppress homologous recombination repair and synergize with PARP inhibitors via regulating SUV39H1 in BRCA-proficient triple-negative breast cancerClin Cancer Res20162271699171226546619
- JudesGRifaïKDauresMDuboisLBignonYJPenault-LlorcaFBernard-GallonDHigh-throughput «Omics» technologies: New tools for the study of triple-negative breast cancerCancer Lett2016 pii: S0304–S3835(16)30137–30139
- ClaudinoWMQuattroneABiganzoliLPestrinMBertiniIDi LeoAMetabolomics: available results, current research projects in breast cancer, and future applicationsJ Clin Oncol200725192840284617502626
- GastMCSchellensJHBeijnenJHClinical proteomics in breast cancer: a reviewBreast Cancer Res Treat20091161172919082706
- MoestueSADamCGGoradSSMetabolic biomarkers for response to PI3K inhibition in basal-like breast cancerBreast Cancer Res2013151R1623448424
- ChenSMGuoCLShiJJHSP90 inhibitor AUY922 abrogates up-regulation of RTKs by mTOR inhibitor AZD8055 and potentiates its antiproliferative activity in human breast cancerInt J Cancer2014135102462247424706460
- LimSOLiCWXiaWEGFR Signaling enhances aerobic glycolysis in triple-negative breast cancer cells to promote tumor growth and immune escapeCancer Res20167651284129626759242
- ZhuXShanLWangFHypermethylation of BRCA1 gene: implication for prognostic biomarker and therapeutic target in sporadic primary triple-negative breast cancerBreast Cancer Res Treat2015150347948625783183
- YoungSPilarskiRTDonenbergTThe prevalence of BRCA1 mutations among young women with triple-negative breast cancerBMC Cancer200998619298662
- NandaRSchummLPCummingsSGenetic testing in an ethnically diverse cohort of high-risk women: A comparative analysis of brca1 and brca2 mutations in american families of european and african ancestryJAMA2005294151925193316234499
- ChenXSYuanYGarfieldDHWuJYHuangOShenKWBoth carboplatin and bevacizumab improve pathological complete remission rate in neoadjuvant treatment of triple negative breast cancer: a meta-analysisPLoS One201499e10840525247558
- EggerGLiangGAparicioAJonesPAEpigenetics in human disease and prospects for epigenetic therapyNature2004429699045746315164071
- JaenischRBirdAEpigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signalsNat Genet200333Suppl24525412610534
- HsuNCHuangYFYokoyamaKKChuPYChenFMHouMFMethylation of BRCA1 promoter region is associated with unfavorable prognosis in women with early-stage breast cancerPLoS One201382e5625623405268
- ToffoliSBarIAbdel-SaterFIdentification by array comparative genomic hybridization of a new amplicon on chromosome 17q highly recurrent in BRCA1 mutated triple negative breast cancerBreast Cancer Res201416646625416589
- XuYDiaoLChenYPromoter methylation of BRCA1 in triple-negative breast cancer predicts sensitivity to adjuvant chemotherapyAnn Oncol20132461498150523406733
- SainiKSSainiMLMarbaixEBiobanking in the era of precision oncologyIndian J Med Paediatr Oncol20153611225810568
- WagnerPDSrivastavaSNew paradigms in translational science research in cancer biomarkersTransl Res2012159434335322424436
- HouthuijzenJMDaenenLGMRoodhartJMLVoestEEThe role of mesenchymal stem cells in anti-cancer drug resistance and tumour progressionBr J Cancer2012106121901190622596239
- XinYLiJWuJPharmacokinetic and pharmacodynamic analysis of circulating biomarkers of anti-NRP1, a novel antiangiogenesis agent, in two phase I trials in patients with advanced solid tumorsClin Cancer Res201218216040604822962439
- AchenMGStackerSAThe vascular endothelial growth factor family; proteins which guide the development of the vasculatureInt J Exp Pathol199879525526510193309
- NishidaNYanoHNishidaTKamuraTKojiroMAngiogenesis in cancerVasc Health Risk Manag20062321321917326328
- NingQLiuCHouLVascular endothelial growth factor receptor-1 activation promotes migration and invasion of breast cancer cells through epithelial-mesenchymal transitionPLoS One201386e6521723776453
- RobertiMPArriagaJMBianchiniMProtein expression changes during human triple negative breast cancer cell line progression to lymph node metastasis in a xenografted model in nude miceCancer Biol Ther201213111123114022825326
- BahhnassyAMohanadMShaarawySTransforming growth factor-β, insulin-like growth factor I/insulin-like growth factor I receptor and vascular endothelial growth factor-A: prognostic and predictive markers in triple-negative and non-triple-negative breast cancerMol Med Rep201512185186425824321
- GhoshSSullivanCAZerkowskiMPHigh levels of vascular endothelial growth factor and its receptors (VEGFR-1, VEGFR-2, neuropilin-1) are associated with worse outcome in breast cancerHuman Pathol200839121835184318715621
- TahaFMZeeneldinAAHelalAMPrognostic value of serum vascular endothelial growth factor in Egyptian females with metastatic triple negative breast cancerClin Biochem20094213–141420142619576877
- BearHDTangGRastogiPBevacizumab added to neoadjuvant chemotherapy for breast cancerN Engl J Med2012366431032022276821
- TolaneySMBoucherYDudaDGRole of vascular density and normalization in response to neoadjuvant bevacizumab and chemotherapy in breast cancer patientsProc Natl Acad Sci U S A201511246143251433026578779
- AndréFZielinskiCCOptimal strategies for the treatment of metastatic triple-negative breast cancer with currently approved agentsAnn Oncol201223Suppl 6vi46vi5123012302
- NatarajanKXieYBaerMRRossDDRole of breast cancer resistance protein (BCRP/ABCG2) in cancer drug resistanceBiochem Pharmacol20128381084110322248732
- FinnRSBengalaCIbrahimNDasatinib as a single agent in triple-negative breast cancer: results of an open-label phase 2 studyClin Cancer Res201117216905691322028489
- Komeili-MovahhedTFouladdelSBarzegarEPI3K/Akt inhibition and down-regulation of BCRP re-sensitize MCF7 breast cancer cell line to mitoxantrone chemotherapyIran J Basic Med Sci201518547247726124933
- VillareteLHRemickDGTranscriptional and post-transcriptional regulation of interleukin-8Am J Pathol19961495168516938909257
- MonteroJCEsparis-OgandoARe-LouhauMFActive kinase profiling, genetic and pharmacological data define mTOR as an important common target in triple-negative breast cancerOncogene201433214815623246963
- BaxiSMTanWMurphySTSmealTYinMJTargeting 3-phos-phoinoside - dependent kinase-1 to inhibit insulin-like growth factor-I induced AKT and p70 S6 kinase activation in breast cancer cellsPLoS One2012710e4840223119004
- EdlingCEHallbergBc-Kit – a hematopoietic cell essential receptor tyrosine kinaseInt J Biochem Cell Biol200739111995199817350321
- NogiHKobayashiTSuzukiMEGFR as paradoxical predictor of chemosensitivity and outcome among triple-negative breast cancerOncol Rep200921241341719148516
- CheangMCVoducDBajdikCBasal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotypeClin Cancer Res20081451368137618316557
- CareyLARugoHSMarcomPKTBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancerJ Clin Oncol201230212615262322665533
- ToyamaTYamashitaHKondoNFrequently increased epidermal growth factor receptor (EGFR) copy numbers and decreased BRCA1 mRNA expression in Japanese triple-negative breast cancersBMC Cancer2008830918950515
- TengYHTanWJThikeAAMutations in the epidermal growth factor receptor (EGFR) gene in triple negative breast cancer: possible implications for targeted therapyBreast Cancer Res2011132R3521457545
- JacotWLopez-CrapezEThezenasSLack of EGFR-activating mutations in European patients with triple-negative breast cancer could emphasise geographic and ethnic variations in breast cancer mutation profilesBreast Cancer Res201113619
- MarzecKABaxterRCMartinJLTargeting insulin-like growth factor binding protein-3 signaling in triple-negative breast cancerBioMed Res Int2015201563852626221601
- ZhaJLacknerMRTargeting the insulin-like growth factor receptor-1R pathway for cancer therapyClin Cancer Res20101692512251720388853
- WangCGaoCMengKQiaoHWangYHuman adipocytes stimulate invasion of breast cancer MCF-7 cells by secreting IGFBP-2PLoS One2015103e011934825747684
- RoseDPHaffnerSMBaillargeonJAdiposity, the metabolic syndrome, and breast cancer in African-American and white American womenEndocr Rev200728776377717981890
- MountziosGAivaziDKostopoulosIDifferential expression of the insulin-like growth factor receptor among early breast cancer subtypesPLoS One201493e9140724637962
- SohnJDoKALiuSFunctional proteomics characterization of residual triple-negative breast cancer after standard neoadjuvant chemotherapyAnn Oncol201324102522252623925999
- HernandezBYWilkensLRLe MarchandLHorioDChongCDLooLWDifferences in IGF-axis protein expression and survival among multiethnic breast cancer patientsCancer Med20154335436225619494
- MartinJLde SilvaHCLinMZScottCDBaxterRCInhibition of insulin-like growth factor-binding protein-3 signaling through sphingosine kinase-1 sensitizes triple-negative breast cancer cells to EGF receptor blockadeMol Cancer Ther201413231632824337110
- LiJSongZWangYOverexpression of SphK1 enhances cell proliferation and invasion in triple-negative breast cancer via the PI3K/AKT signaling pathwayTumour Biol2016378105871059326857281
- ThikeAACheokPYJara-LazaroARTanBTanPTanPHTriple-negative breast cancer: clinicopathological characteristics and relationship with basal-like breast cancerMod Pathol201023112313319855377
- ZhuYWangYGuanBC-kit and PDGFRA gene mutations in triplenegative breast cancerInt J Clin Exp Pathol2014774280428525120810
- JohanssonIAaltonenKEEbbessonAIncreased gene copy number of KIT and VEGFR2 at 4q12 in primary breast cancer is related to an aggressive phenotype and impaired prognosisGenes Chromosomes Cancer201251437538322170730
- JanssonSBendahl PäOGrabauDAThe three receptor tyrosine kinases c-KIT, VEGFR2 and PDGFRα, closely spaced at 4q12, show increased protein expression in triple-negative breast cancerPLoS One201497e10217625025175
- CuriglianoGPivotXCortesJRandomized phase II study of sunitinib versus standard of care for patients with previously treated advanced triple-negative breast cancerBreast201322565065623958375
- OrganSLTsaoMSAn overview of the c-MET signaling pathwayTher Adv Med Oncol201131 SupplS7S1922128289
- HsuYHYaoJChanLCDefinition of PKC-alpha, CDK6, and MET as therapeutic targets in triple-negative breast cancerCancer Res201474174822483524970481
- Gonzalez-AnguloAMChenHKaruturiMSFrequency of mesenchymal-epithelial transition factor gene (MET) and the catalytic subunit of phosphoinositide-3-kinase (PIK3CA) copy number elevation and correlation with outcome in patients with early stage breast cancerCancer2013119171522736407
- YiYWYouKBaeEJKwakSJSeongYSBaeIDual inhibition of EGFR and MET induces synthetic lethality in triple-negative breast cancer cells through downregulation of ribosomal protein S6Int J Oncol201547112213225955731
- DierasVCamponeMYardleyDARandomized, phase II, placebo-controlled trial of onartuzumab and/or bevacizumab in combination with weekly paclitaxel in patients with metastatic triple-negative breast cancerAnn Oncol20152691904191026202594
- TolaneySMTanSGuoHPhase II study of tivantinib (ARQ 197) in patients with metastatic triple-negative breast cancerInvest New Drugs20153351108111426123926
- YanSJiaoXZouHLiKPrognostic significance of c-Met in breast cancer: a meta-analysis of 6010 casesDiagn Pathol2015106226047809
- SolimanHKhalilFAntoniaSPD-L1 expression is increased in a subset of basal type breast cancer cellsPLoS One201492e8855724551119
- MuenstSSchaerliARGaoFExpression of programmed death ligand 1 (PD-L1) is associated with poor prognosis in human breast cancerBreast Cancer Res Treat20141461152424842267
- GatalicaZSnyderCManeyTProgrammed cell death 1 (PD-1) and its ligand (PD-L1) in common cancers and their correlation with molecular cancer typeCancer Epidemiol Biomarkers Prev201423122965297025392179
- MittendorfEAPhilipsAVMeric-BernstamFPD-L1 Expression in triple negative breast cancerCancer Immunol Res20142436137024764583
- NandaRChowLQDeesECPembrolizumab in patients with advanced triple-negative breast cancer: phase Ib KEYNOTE-012 studyJ Clin Oncol201634212460246727138582
- HollanderMCBlumenthalGMDennisPAPTEN loss in the continuum of common cancers, rare syndromes and mouse modelsNat Rev Cancer201111428930121430697
- KeirMEButteMJFreemanGJSharpeAHPD-1 and its ligands in tolerance and immunityAnnu Rev Immunol20082667770418173375
- PandeMBondyMLDoKAAssociation between germline single nucleotide polymorphisms in the PI3K-AKT-mTOR pathway, obesity, and breast cancer disease-free survivalBreast Cancer Res Treat2014147238138725108739
- MillerTWBalkoJMArteagaCLPhosphatidylinositol 3-kinase and antiestrogen resistance in breast cancerJ Clin Oncol201129334452446122010023
- TaneiTMorimotoKShimazuKAssociation of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential paclitaxel and epirubicin-based chemotherapy for breast cancersClin Cancer Res200915124234424119509181
- PowellEShaoJYuanYp53 deficiency linked to B cell translocation gene 2 (BTG2) loss enhances metastatic potential by promoting tumor growth in primary and metastatic sites in patient-derived xenograft (PDX) models of triple-negative breast cancerBreast Cancer Res20161811326818199
- ShahSPRothAGoyaRThe clonal and mutational evolution spectrum of primary triple negative breast cancersNature2012486740339539922495314
- CantleyLCThe Phosphoinositide 3-kinase pathwayScience200229655731655165712040186
- YamnikRLDigilovaADavisDCBrodtZNMurphyCJHolzMKS6 kinase 1 regulates estrogen receptor alpha in control of breast cancer cell proliferationJ Biol Chem2009284106361636919112174
- GingrasACGygiSPRaughtBRegulation of 4E-BP1 phosphorylation: a novel two-step mechanismGenes Dev199913111422143710364159
- BraunsteinSKarpishevaKPolaCA hypoxia-controlled capdependent to cap-independent translation switch in breast cancerMol Cell200728350151217996713
- ZhouXTanMStone HawthorneVActivation of the Akt/mammalian target of rapamycin/4E-BP1 pathway by ErbB2 overexpression predicts tumor progression in breast cancersClin Cancer Res200410206779678815501954
- ShrivastavaSKulkarniPThummuriDPiperlongumine, an alkaloid causes inhibition of PI3 K/Akt/mTOR signaling axis to induce caspase-dependent apoptosis in human triple-negative breast cancer cellsApoptosis20141971148116424729100
- DePSunYCarlsonJHFriedmanLSLeyland-JonesBRDeyNDoubling down on the PI3K-AKT-mTOR pathway enhances the antitumor efficacy of PARP inhibitor in triple negative breast cancer model beyond BRCA-nessNeoplasia2014161437224563619
- WalshSFlanaganLQuinnCmTOR in breast cancer: differential expression in triple-negative and non-triple-negative tumorsBreast201221217818221963359
- AlabertCChromatin dynamics during DNA replication and uncharacterized replication factors determined by nascent chromatin capture (NCC) proteomicsNat Cell Biol201416328129324561620
- AlagozMKatsukiYOgiwaraHSETDB1, HP1 and SUV39 promote repositioning of 53BP1 to extend resection during homologous recombination in G2 cellsNucleic Acids Research201543167931794426206670
- ChoiJDParkMALeeJSSuppression and recovery of BRCA1-mediated transcription by HP1gamma via modulation of promoter occupancyNucleic Acids Res20124022113211133823074186
- TanMHMesterJLNgeowJRybickiLAOrloffMSEngCLifetime cancer risks in individuals with germline PTEN mutationsClin Cancer Res201218240040722252256
- HicksCKumarRPannutiAAn integrative genomics approach for associating GWAS information with triple-negative breast cancerCancer Inform20131212023423317
- BegSSirajAKPrabhakaranSLoss of PTEN expression is associated with aggressive behavior and poor prognosis in Middle Eastern triple-negative breast cancerBreast Cancer Res Treat2015151354155325981902
- GrindeMTSkrboNMoestueSAInterplay of choline metabolites and genes in patient-derived breast cancer xenograftsBreast Cancer Res2014161R524447408
- TomitaHTanakaKTanakaTHaraAAldehyde dehydrogenase 1A1 in stem cells and cancerOncotarget2016710110181103226783961
- KimSJKimYSJangEDSeoKJKimJSPrognostic impact and clinicopathological correlation of CD133 and ALDH1 expression in invasive breast cancerJ Breast Cancer201518434735526770241
- MarcatoPDeanCAPanDAldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasisStem Cells2011291324521280157
- OhiYUmekitaYYoshiokaTAldehyde dehydrogenase 1 expression predicts poor prognosis in triple-negative breast cancerHistopathology201159477678022014057
- ProctorEKidwellKMJiaggeECharacterizing breast cancer in a population with increased prevalence of triple-negative breast cancer: androgen receptor and ALDH1 expression in ghanaian womenAnn Surg Oncol201522123831383525743329
- LehmannBDBauerJAChenXSandersMEChakravarthyABShyrYPietenpolJAIdentification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapiesJ Clin Invest201112172750276721633166
- ShiovitzSKordeLAGenetics of breast cancer: a topic in evolutionAnn Oncol20152671291129925605744
- UrruticoecheaASmithIEDowsettMProliferation marker Ki-67 in early breast cancerJ Clin Oncol200523287212722016192605
- TungNLinNUKiddJFrequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancerJ Clin Oncol201634131460146826976419
- TurnerNTuttAAshworthAHallmarks of ‘BRCAness’ in sporadic cancersNat Rev Cancer200441081481915510162
- LeeLJAlexanderBSchnittSJComanderAGallagherBGarberJETungNClinical outcome of triple negative breast cancer in BRCA1 mutation carriers and non-carriersCancer2011117143093310021264845
- DentRTrudeauMPritchardKITriple-negative breast cancer: clinical features and patterns of recurrenceClin Cancer Res200713154429443417671126
- FoulkesWDMetcalfeKHannaWDisruption of the expected positive correlation between breast tumor size and lymph node status in BRCA1-related breast carcinomaCancer20039881569157714534871
- AndersonDEFamilial versus sporadic breast cancerCancer1992706 Suppl174017461516029
- AtchleyDPAlbarracinCTLopezAClinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancerJ Clin Oncol200826264282428818779615
- MusolinoABellaMABortesiBBRCA mutations, molecular markers, and clinical variables in early-onset breast cancer: a population-based studyBreast200716328029217257844
- StefanssonOAJonassonJGJohannssonOTOlafsdottirKSteinarsdottirMValgeirsdottirSEyfjordJEGenomic profiling of breast tumours in relation to BRCA abnormalities and phenotypesBreast Cancer Res2009114R4719589159
- ZugazagoitiaJPérez-SeguraPManzanoALimited family structure and triple-negative breast cancer (TNBC) subtype as predictors of BRCA mutations in a genetic counseling cohort of early-onset sporadic breast cancersBreast Cancer Res Treat2014148241542125342642
- StefanssonOAJonassonJGOlafsdottirKCpG island hypermethylation of BRCA1 and loss of pRb as co-occurring events in basal/triple-negative breast cancerEpigenetics20116563864921593597
- BranhamMTMarzeseDMLauritoSRMethylation profile of triple-negative breast carcinomasOncogenesis201217e1723552734
- FoedermayrMSebestaMRudasMBRCA-1 methylation and TP53 mutation in triple-negative breast cancer patients without pathological complete response to taxane-based neoadjuvant chemotherapyCancer Chemother Pharmacol201473477177824526178
- Mitre-AguilarIBCabrera-QuinteroAJZentella-DehesaAGenomic and non-genomic effects of glucocorticoids: implications for breast cancerInt J Clin Exp Pathol20158111025755688
- VilascoMCommunalLHugon-RodinJLoss of glucocorticoid receptor activation is a hallmark of BRCA1-mutated breast tissueBreast Cancer Res Treat2013142228329624166279
- ReederAAttarMNazarioLStress hormones reduce the efficacy of paclitaxel in triple negative breast cancer through induction of DNA damageBr J Cancer201511291461147025880007
- SkorMNWonderELKocherginskyMGoyalAHallBACaiYConzenSDGlucocorticoid receptor antagonism as a novel therapy for triple-negative breast cancerClin Cancer Res201319226163617224016618
- AntonovaLMuellerCRHydrocortisone down-regulates the tumor suppressor gene BRCA1 in mammary cells: a possible molecular link between stress and breast cancerGenes Chromosomes Cancer200847434135218196591
- RitterHDAntonovaLMuellerCRThe unliganded glucocorticoid receptor positively regulates the tumor suppressor gene BRCA1 through GABP betaMol Cancer Res201210455856922328717
- BirchJMAlstonRDMcNallyRJRelative frequency and morphology of cancers in carriers of germline TP53 mutationsOncogene200120344621462811498785
- KimYKimJLeeHDJeongJLeeWLeeKASpectrum of EGFR gene copy number changes and KRAS gene mutation status in korean triple negative breast cancer patientsPLoS One2013810e7901424205362
- KimJYParkKJungHHAssociation between mutation and expression of TP53 as a potential prognostic marker of triple-negative breast cancerCancer Res Treat Epub2016218
- ÇakırTKhatibipourMJMetabolic network discovery by top-down and bottom-up approaches and paths for reconciliationFront Bioeng Biotechnol201426225520953
- DeanSJPerksCMHollyJMLoss of PTEN expression is associated with IGFBP2 expression, younger age, and late stage in triple-negative breast cancerAm J Clin Pathol2014141332333324515759
