Abstract
Background
O6-methylguanine-DNA methyltransferase (MGMT) is an independent predictor of therapeutic response and potential prognosis in patients with glioblastoma multiforme (GBM). However, its significance of clinical prognosis in different continents still needs to be explored.
Patients and methods
To explore the effects of MGMT promoter methylation on both progression-free survival (PFS) and overall survival (OS) among GBM patients from different continents, a systematic review of published studies was conducted.
Results
A total of 5103 patients from 53 studies were involved in the systematic review and the total percentage of MGMT promoter methylation was 45.53%. Of these studies, 16 studies performed univariate analyses and 17 performed multivariate analyses of MGMT promoter methylation on PFS. The pooled hazard ratio (HR) estimated for PFS was 0.55 (95% CI 0.50, 0.60) by univariate analysis and 0.43 (95% CI 0.38, 0.48) by multivariate analysis. The effect of MGMT promoter methylation on OS was explored in 30 studies by univariate analysis and in 30 studies by multivariate analysis. The combined HR was 0.48 (95% CI 0.44, 0.52) and 0.42 (95% CI 0.38, 0.45), respectively.
Conclusion
In each subgroup divided by areas, the prognostic significance still remained highly significant. The proportion of methylation in each group was in inverse proportion to the corresponding HR in the univariate and multivariate analyses of PFS. However, from the perspective of OS, compared with data from Europe and the US, higher methylation rates in Asia did not bring better returns.
Introduction
Glioblastoma multiforme (GBM, WHO grade 4) is the most common primary brain tumor in adults with an annual incidence of 3–4/100,000 and is associated with poor prognosis.Citation1 Although some clinical trials have demonstrated that the standard treatment improves overall survival (OS) and progression-free survival (PFS), only less than one-third of GBM patients seem to benefit from these therapies, mainly because of GBM resistance to alkylating drugs.
Transcriptionally active O6-methylguanine-DNA methyltransferase (MGMT) gene encodes a ubiquitously expressed suicide DNA repair enzyme that counteracts the normally lethal effects of alkylating agents by removing the alkyl adducts, preventing the formation of cross-links and thereby causing resistance to alkylating drugs.Citation3 The loss of MGMT protein expression caused by methylation of the MGMT promoter reduces the DNA repair activity of glioma cells, preventing their resistance to alkylating agents.Citation2,Citation4–Citation6 It is believed that patients with GBM who have a methylated MGMT promoter are more sensitive to the killing effects of alkylating drugs, because tumor cells with low MGMT expression were unable to repair such DNA lesions and, thus, were prone to apoptosis, whereas those that do not have a methylated MGMT promoter do not have this benefit.Citation68,Citation69
Various studies have shown that the MGMT promoter methylation status is an independent prognostic factor to GBM and the assessment of MGMT promoter methylation is currently considered as mandatory for patient selection in clinical trials.Citation7–Citation10,Citation68 However, many differences in high risk factors and postoperative chemoradiation stay in guidelines for the treatment of glioblastoma, among countries, indicating different attitudes to MGMT promoter methylation status. Is the prognostic significance of MGMT promoter methylation independent equally among glioblastomas from different areas? Further explorations are needed in the prognostic value of MGMT promoter methylation on GBM including therapeutic intervention.Citation11,Citation12, Citation20, Citation21
From the perspective of geography, we conducted this meta-analysis to test the independence of prognostic value of MGMT promoter methylation in both PFS and OS among patients with GBM.
Patients and methods
Publication selection
Ethical approval and patient consent are not required as this is a systematic review and meta-analysis of previously published studies. This study was performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement.Citation13
Two reviewers (Yangyang Jiang and Wei Meng) participated in the citations search, study selection and data extraction, independently. Divergences between reviewers were resolved through consulting with Professor Jie Ma.
Electronic databases, including PubMed, EMBASE, Web of Science, China Biomedical Literature Database (CBM), Chinese National Knowledge Infrastructure (CNKI), China Wan Fang database and the Cochrane library, were searched for relevant clinical trials published on the association between MGMT promoter methylation and GBM between January 2000 and June 2017.
The search combined key words: (“O6-methylguanine-DNA methyltransferase methylation” OR MGMT methylation”) AND (“glioblastoma” OR “GBM”) AND (“survival analysis” OR “meta analysis”) AND (“MSP” OR “PSQ”) AND (“survival analysis” OR “meta analysis”) AND (“methylation-specific polymerase chain reaction and pyrosequencing”).
The meta-analysis gathered complete databases from published cohort studies dealing with the prognostic value of MGMT promoter methylation in patients with GBM no matter which therapy was given.
The language in which the papers were written was restricted to English and Chinese. Abstracts were excluded because of insufficient data for meta-analysis. In order to identify the relevant publications, the references cited in the research papers were also scanned. To avoid duplication of data, we carefully noted the author names and the different research centers involved. We evaluated the eligible studies if all the following conditions were met: 1) MGMT promoter methylation status was measured by using identified method such as methylation-specific polymerase chain reaction (MSP) and pyrosequencing (PSQ); (2) inclusion of sufficient data or survival curves to calculate hazard ratio (HR) and 95% CI; and 3) full or special parts of papers investigated the relationship between MGMT promoter methylation and PFS or OS.
Data extraction
Two authors (Yangyang Jiang and Wei Meng) independently reviewed and extracted the data needed. Disagreements were resolved through discussion with each other.
We used a predesigned data extraction sheet to obtain the following information: first author, year of publication, region, HR form and sample size and style of postoperative chemoradiation, if given. The formula recommended by Spruance et al was adopted to calculate the corresponding HR of the missing data.Citation14 Kaplan–Meier curve was read by using Engauge Digitizer version 4.1 (available at: http://source-forge.net/) except if the paper has supplied HR directly.Citation15 (All the data are shown in .)
Table 1 Main characteristics and results of eligible studies
Statistical analysis
In some studies, HR and 95% CI were directly obtained from published literature by using univariate or multivariate survival analysis. For studies in which the HR corresponding to the 95% CI was not given directly, published data and figures from original papers were used to calculate the HR according to the methods described by using Engauge Digitizer version 4.1.
The pooled HR corresponding to the 95% CI was used to assess the prognostic value of MGMT promoter methylation in patients with GBM. The statistical heterogeneity among studies was assessed with the Q-test and I2 statistics.Citation16
A random-effects model was used primarily regardless of heterogeneity. Level of heterogeneity (level of variance) across studies was evaluated using I2 statistic. I2 of 40, 70 and 100% was used to represent low, moderate and high variance, respectively.Citation17 If obvious differences for clinical characteristics and methodology were not identified and I2 ≤ 40%, a fixed-effects model was adopted. A random-effects model will be used if clinical characteristics and methodology were not identified to be great difference and I2 ≤ 40%; in contrast, if the clinical characteristic and/or methodology across studies regardless of I2 statistic was considered to be obviously different, qualitative analysis was adopted.Citation18
The objective impact of MGMT promoter methylation on PFS and OS was considered to be statistically significant if the 95% CI for the HR did not overlap 0. Publication bias was evaluated with funnel plot and Begg’s rank correlation method.Citation19 The statistical analyses were performed by STATA/MP 13.0 software.
Results
Characteristics of studies
A total of 204 relevant citations were identified at the initial search stage; 151 articles concerned topics not relevant to this study, and finally 53 studies were included in the meta-analysis.
All the included studies were in English. The individual characteristics of the eligible studies are reported in . The total number of patients was 5103, and the total frequency of MGMT promoter methylation was 45.53%. Of the 53 publications eligible for systematic review, 31 studies reported the HR with corresponding to 95% CI directly, the other 22 studies reported the HR in the style of survival curve availability.
Meta-analysis
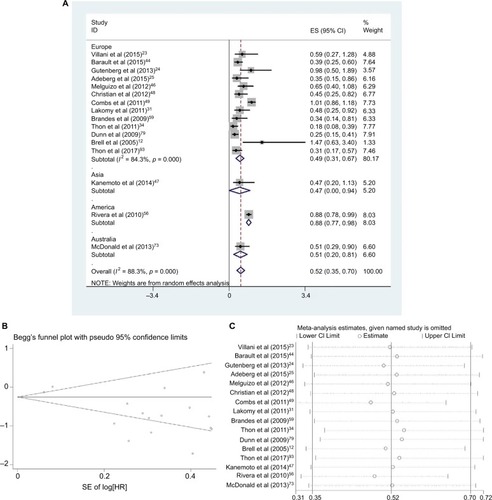
Sixteen studies (one in Asia, one in North America, one in Australia and 13 in Europe) reported the effect of MGMT promoter methylation on PFS using univariate analysis.Citation12,Citation23–Citation25,Citation31,Citation44,Citation46–Citation49,Citation56,Citation59,Citation68,Citation73,Citation79,Citation93 As shown in , the HR of the Asian group is 0.47, the HR of the American group is 0.88, the HR of the Australian group is 0.51 and the HR of the European group is 0.49; MGMT promoter methylation was significantly correlated with better PFS according to univariate analysis, with a combined HR of 0.55 (95% CI 0.50, 0.60). The random-effects model (the DerSimonian and Laird method) was used because significant heterogeneity was detected among these studies (p = 0.000, I2 = 88.3%).Citation61
Figure 1 Data statistics on PFS using univariate analysis.
Abbreviations: SE, standard error; ES, effect size; HR, hazard ratio; MGMT, O6-methylguanine-DNA methyltransferase; PFS, progression-free survival.
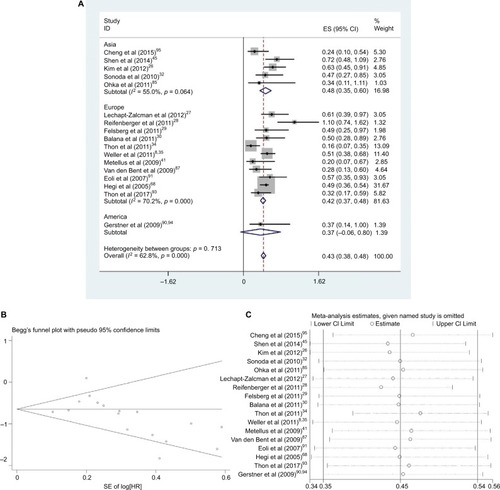
The effect of MGMT promoter methylation on PFS adjusted for other variables was evaluated in 17 studies (five in Asia, 11 in Europe and one in America)Citation26–Citation30,Citation32,Citation34,Citation35,Citation41,Citation45,Citation68,Citation85,Citation87,Citation91,Citation93–Citation95 As shown in , the HR of the Asian group is 0.49, the HR of the European group is 0.44 and the HR of the American group is 0.37; MGMT promoter methylation was significantly correlated with better PFS according to multivariate analysis, with a combined HR of 0.45 (95% CI 0.35, 0.54). The random-effects model (the DerSimonian and Laird method) was used because significant heterogeneity was detected among these studies (p = 0.000, I2 = 62.8%).Citation61
Figure 2 Data statistics on PFS using multivariate analysis.
Abbreviations: SE, standard error; ES, effect size; HR, hazard ratio; MGMT, O6-methylguanine-DNA methyltransferase; PFS, progression-free survival.
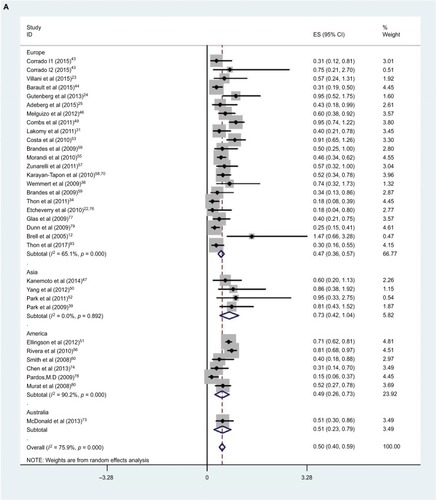
The effect of MGMT promoter methylation on OS unadjusted for using univariate analysis was evaluated in 32 studies (four in Asia, six in North America, one in Australia and 21 in Europe).Citation12,Citation22–Citation25,Citation29,Citation31,Citation34,Citation36,Citation39,Citation43,Citation44,Citation46,Citation47,Citation49–Citation60,Citation73,Citation74,Citation77–Citation80,Citation93 As shown in , the HR of the American group is 0.49, the HR of the European group is 0.47, HR of the Asian group is 0.73 and the HR of the Australian group is 0.51; MGMT promoter methylation was significantly correlated with better OS according to univariate analysis, with a combined HR of 0.50 (95% CI 0.40, 0.59). The random-effects model (the DerSimonian and Laird method) was used as significant heterogeneity was detected among these studies (p = 0.000, I2 = 50.3%).Citation61
Figure 3 Data statistics on OS using univariate analysis.
Abbreviations: SE, standard error, ES, effect size; HR, hazard ratio; MGMT, O6-methylguanine-DNA methyltransferase; OS, overall survival.
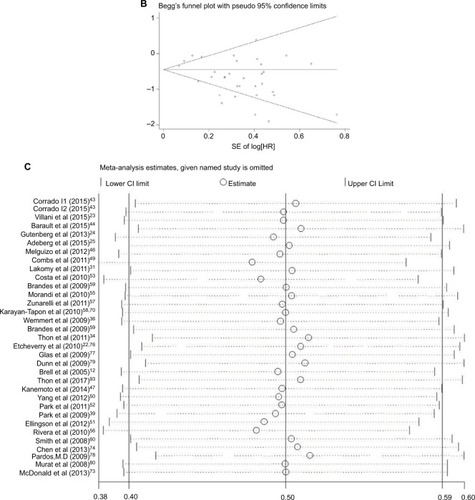
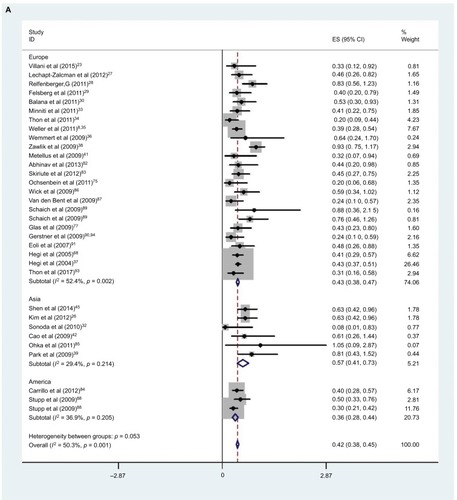
Thirty-one studies (six in Asia, two in America and 23 in Europe) reported the effect of MGMT promoter methylation on OS using analyses adjusted for other factors.Citation23,Citation26–Citation30,Citation33–Citation42,Citation45,Citation68,Citation75,Citation77,Citation82–Citation91,Citation93 As shown in , the HR of the Asian group is 0.56, the HR of the American group is 0.37 and the HR of the European group is 0.44; MGMT promoter methylation was significantly correlated with better OS according to multivariate analysis, with a combined HR of 0.44 (95% CI 0.38, 0.50). The random-effects model (the DerSimonian and Laird method) was used as significant heterogeneity was detected among these studies (p = 0.000, I2 = 50.3%).Citation61
Figure 4 Data statistics on OS using multivariate analysis.
Abbreviations: SE, standard error; ES, effect size; HR, hazard ratio; MGMT, O6-methylguanine-DNA methyltransferase; OS, overall survival.
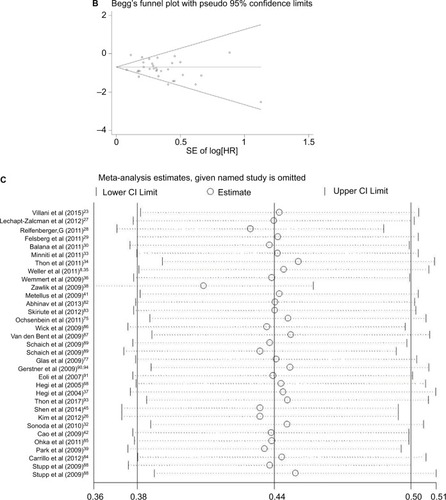
Publication bias statistics were determined; some publication bias (Begg’s test, p<0.05) was found. Sensitivity analysis was performed to investigate the influence of a single study on the overall meta-analysis by omitting one study at a time, and the omission of any study made no significant difference, indicating that our results were statistically reliable.
Discussion
The association between the MGMT promoter methylation and GBM has been reported in many studies. Evaluations of prognostic factors, such as patients age, gender, nationality, recurrence, tumor location and excision, MGMT testing method and the style of postoperative chemoradiation for tumors are all vital to improve research pursuing new therapies for GBM. In general, the population flows more and more frequently among the continents, and most of the prognostic factors are usually determined by circumstances and nationwide medical policies. Therefore, it is more reasonable to set subgroups by areas but not by races. Our meta-analysis was performed to define the prognostic and predictive value of MGMT promoter methylation in glioblastoma patients of different continents. The major strengths of this study include the deliberate distinction of area, the relatively comprehensive sample size, the prospective data collection and the combination of the MSP and PSQ analysis to assess the MGMT promoter methylation status.
MGMT expression protects normal cells from carcinogens; however, it can also protect cancer cells from chemotherapeutic alkylating agents, which include mutations, sister chromatid exchanges, recombination and chromosomal aberrations.Citation62 It has been shown that glial brain tumors are characterized by a low expression of MGMT, however, the activity of MGMT is commonly increased in relation to surrounding normal tissue.Citation63,Citation64
The data of the Adeberg et al study show that delaying postoperative chemoradiation for GBM patients – carried out in order to determine MGMT promoter status – did not have a negative impact on survival time. Indeed, initiating radiation therapy sooner than 24 days after surgery has a negative impact on progression and survival.Citation25
In the older glioblastoma patient, MGMT promoter meth-ylation status is still contentious on clinical decision making. For the elderly with malignant glioma, two recently published Phase III trials have evaluated the place of dose-dense/conventional temozolomide (TMZ) regimes alone as compared with conventional/hypofractionated radiotherapy.Citation65–Citation67 OS in methylated patients was better if TMZ treatment was applied, whereas in unmethylated patients radiotherapy alone was more effective. However, in contrast, Gutenberg et al study showed no significant differences in OS for concomitant plus adjuvant administration of TMZ, as the current standard treatment specifies, to sequentially administered TMZ.Citation24 Concerning age, the findings of Gutenberg et al suggest that patients over 65 years of age showed significantly longer PFS and a trend toward longer OS when receiving concomitant plus adjuvant TMZ as compared to the sequential TMZ regimen. Thus, MGMT promoter methylation is an important biomarker for personalized treatment strategies in the elderly subpopulation.
It was found that GBM patients with MGMT promoter methylation had better OS and PFS than those without methylated status by univariate or multivariate analysis regardless of therapeutic intervention and area.Citation72,Citation74–Citation78 MGMT gene promoter methylation levels can be used as a sensitive biomarker of using alkylating agents in GBM patients.Citation86,Citation88,Citation89,Citation92 The results suggested that MGMT promoter methylation indicated better clinical prognosis of GBM, and played an independent role with GBM development.Citation80,Citation82–Citation84 Yang et al once have explored the connection between MGMT promoter methylation in glioblastoma and different race, conducting a primary conclusion, GBM patients with MGMT promoter methylation only had longer OS by multivariate analysis in Asian, but with no further exploration of subgroup.Citation81 Also, because of population flow, it is more reasonable and accurate to set subgroup by continent but not by race. Therefore, is the prognostic significance of MGMT promoter methylation independent equally in glioblastomas of different areas?
In our univariate analysis of PFS, MGMT promoter methylation ratio of Asian groups is 0.67, the European is 0.41 and the American is 0.24. The HR of Asian groups is 0.47, the European is 0.49 and the American is 0.88. The proportion of methylation in each group was in inverse proportion to the corresponding HR. In our multivariate analysis of PFS, MGMT promoter methylation ratio of Asian groups is 0.29, the European is 0.53 and the American is 0.58. The HR of Asian groups is 0.49, the European is 0.44 and the American is 0.37. The proportion of methylation in each group was also in inverse proportion to the corresponding HR.
In our univariate analysis of OS, MGMT promoter methylation ratio of Asian groups is 0.50, the European is 0.46, the Australia is 0.36 and the American is 0.35. The HR of Asian groups is 0.73, the European is 0.47, the Australia is 0.51 and the American is 0.49. The proportion of methylation in most groups was in inverse proportion to the corresponding HR except for the Asian group. In our multivariate analysis of OS, MGMT promoter methylation ratio of Asian groups is 0.53, the European is 0.53 and the American is 0.72. The HR of Asian groups is 0.56, the European is 0.43 and the American is 0.36. The proportion of methylation in the European and American group was in inverse proportion to the corresponding HR but Asian group doesn’t follow the inverse relation.
Our meta-analysis with pooled data suggested that MGMT promoter methylation was associated with prolonged PFS in GBM patients according to both univariate analysis and multivariate analysis. From the perspective of PFS, the prognostic significance of MGMT promoter methylation is independent and basically equal in glioblastomas of different areas. Prolonged OS in GBM patients was also accompanied by MGMT promoter methylation through univariate analysis and multivariate analysis. However, from the perspective of OS, the prognostic significance of MGMT promoter methylation in the Asian group was not so important as in the European and American groups.
There are still two public questions. First, what is the most appropriate method for the assessment of methylation? The various technologies of measurement of MGMT promoter methylation sometimes show discrepant or even opposite results. It is originally regarded that MSP which evaluates the methylation status of the MGMT promoter is the best way to predict the MGMT expression of the tumor in a manner that also correlates with clinical prognosis.Citation56 In the last 5 years, more and more studies have reported that a series of more accurate values have been obtained by PSQ compared to MSP.Citation70 Studies with PSQ showed that this technique, having a higher reproducibility and sensitivity than MSP, is also a qualitative method. Therefore, besides MSP, our meta-analysis also absorbed measurement of MGMT promoter methylation from PSQ, which make our results more persuasive.
Second, what is the best threshold indicating methylated or unmethylated status? The definition of a prognostically relevant threshold for the percentage of MGMT methylation remains one of the most critical issues in the use of PSQ analysis. In 2015, the Receiver Operating Characteristics analysis from Villani et al showed that the best possible criteria for PSQ-detected percentage of MGMT methylation that predicted PFS and OS were 19% and 13%, respectively.Citation23
This meta-analysis has several potential limitations that may be taken into account. First, only English and Chinese language literature studies were scanned for publication. If the search had included literature studies published in other languages, it is possible that more additional relevant trials may have been considered. Second, some ongoing studies, most of which being of high quality, were ineligible for inclusion. Therefore, limitations in quality cannot be excluded, and the pooled results of this meta-analysis may have been affected, more or less. Moreover, subgroup analysis still needs a larger number of trials to make results convincing. Additionally, we are unable to assess the effects of other clinically meaningful end points on PFS or OS, such as quality of life, patient and physician satisfaction with surgical resection and cytotoxic chemotherapy with the alkylating agent TMZ or concomitant radiotherapy, because of sparse and inconsistent reporting across studies. Finally, because all of the Asian studies included in the meta-analysis were carried out in Japan and South Korea, clinicians and pharmacists should carefully and judiciously assess the feasibility of applying the results in the clinical setting in China.
Conclusion
MGMT promoter methylation was an independent indicator of better prognosis for GBM and epigenetic MGMT gene silencing by promoter methylation associated with loss of MGMT expression may contribute to diminished DNA repair, which may be the potential mechanism that results in longer PFS and OS.Citation71 From the perspective of PFS, the prognostic significance of MGMT promoter methylation is independent and basically equal in glioblastomas of different areas. However, from the perspective of OS, the proportion of methylation in the Asian group was not in basically inverse proportion to the corresponding HR as in European and American groups, in the univariate and multivariate analyses. The different prognosis might result from the intervention of age, percentage of MGMT methylation and the style of postoperative chemoradiation. More exploration is needed to investigate the clinical chemotherapy effect on MGMT promoter of the glioblastoma, screen a more sensitive alkylating agent combination for glioblastoma and apparent genetic targets for potential therapeutic value.
Acknowledgments
The authors thank the Intensive Care Unit of Shanghai Deji Hospital, the Ninth Clinical Medical College of Qingdao University for their help on this article. Our paper has no funding.
Author contributions
Yangyang Jiang and Wei Meng independently reviewed and extracted the data needed. Disagreements were resolved through discussion with Professor Jie Ma. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interests in this work.
References
- StuppRMasonWPvan den BentMJRadiotherapy plus concomitant and adjuv -ant temozolomide for glioblastomaN Engl J Med20053521098799615758009
- HegiMELiuLHermanJGCorrelation of O6-methylguanine methyl-transferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activityJ Clin Oncol200826254189419918757334
- VerbeekBSouthgateTDGilhamDEMargisonGPO6-Methylguanine- DNA methyltransferase inactivation and chemotherapyBr Med Bull200885173318245773
- GersonSLClinical relevance of MGMT in the treatment of cancerJ Clin Oncol20022092388239911981013
- EstellerMGarcia-FoncillasJAndionEInactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agentsN Engl J Med2000343191350135411070098
- BobolaMSBergerMSEllenbogenRGRobertsTSGeyerJRSilberJRO6-Methylguanine-DNA methyltransferase in pediatric primary brain tumors: relation to patient and tumor characteristicsClin Cancer Res20017361311297257
- FriedmanHSMcLendonREKerbyTDNA mismatch repair and O6-alkylguanine-DNA alkyltransferase analysis and response to Temodal in newly diagnosed malignant gliomaJ Clin Oncol19981612385138579850030
- WellerMStuppRReifenbergerGMGMT promoter methylation in malignant gliomas: ready for personalized medicine?Nat Rev Neurol200961395119997073
- ShilpaVBhagatRPremalataCSPallaviVRRameshGKrishnamoorthyLRelationship between promoter methylation & tissue expression of MGMT gene in ovarian cancerIndian J Med Res2014140561662325579142
- FelsbergJRappMLoeserSPrognostic significance of molecular markers and extent of resection in primary glioblastoma patientsClin Cancer Res200915216683669319861461
- SciuscioDDiserensDvan DommelenDMartinetDJonesDJanzerDExtent and patterns of MGMT promoter methylation in glioblastoma-and respective glioblastoma-derived spheresClinical Cancer Research An Official Journal of the American Association for Cancer Research20111725526621097691
- BrellMTortosaAVergerEPrognostic significance of O6-methylguanine-DNA methyl-transferase determined by promoter hypermethylation and immunohistochemical expression in anaplastic gliomasClin Cancer Res200511145167517416033832
- GorliaTvan den BentMJHegiMENomograms for predicting survival of patients with newly diagnosed glioblastoma: prognostic factor analysis of EORTC and NCIC trial 26981–22981/CE.3Lancet Oncol200891293818082451
- SpruanceSLReidJEGraceMSamoreMHazard ratioin clinical trialsAntimicrob Agents Chemother20044882787279215273082
- TierneyJFStewartLAGhersiDPractical methods for incorporating summary time-to-event data into metaanalysisTrials2007811617555582
- CohenJFChalumeauMCohenRKorevaarDAKhoshnoodBBossuytPMCochran’s Q test was useful to assess heterogeneity in likelihood ratios instudies of diagnostic accuracyJ Clin Epidemiol201568329930625441698
- DeeksJJAltmanDGChapter 9 analysing data and undertaking meta-analysesCochrane Handbook for Systematic Reviews of Interventions Version 5.1.02011 Available from: http://xueshu.baidu.com/s?wd=paperuri%3A%2893155479b8a8f47eb878935af3cf9292%29&filter=sc_long_sign&tn=SE_xueshusource_2kduw22v&sc_vurl=http%3A%2F%2Fonlinelibrary.wiley.com%2Fresolve%2Freference%2FXREF%3Fid%3D10.1002%2F9780470712184.ch9&ie=utf-8&sc_us=13279804406616589552Accessed August 29, 2017
- HigginsJPThompsonSGQuantifying heterogeneity in a metaanalysisStat Med200221111539155812111919
- BeggCBMazumdarMOperating characteristics of a rank correlation test for publication biasBiometrics1994504108811017786990
- TangKJinQYanWClinical correlation of MGMT protein expression and promoter methylation in Chinese glioblastoma patientsMed Oncol20122921292129621394635
- MotomuraKNatsumeAKishidaYBenefits of interferon-beta and temozolomide combination therapy for newly diagnosed primary glioblastoma with the unmethylated MGMT promoter: a multicenter studyCancer201111781721173021472719
- EtcheverryAAubryMde TayracMDNA methylation in glioblastoma: impact on gene expression and clinical outcomeBMC Genomics201011170121156036
- VillaniVCasiniBPaceAThe prognostic value of pyrosequencing-detected MGMT promoter hypermethylation in newly diagnosed patients with glioblastomaDis Markers2015201560471926347581
- GutenbergABockHCReifenbergerGBrückWGieseAToxicity and survival in primary glioblastoma patients treated with concomitant plus adjuvant temozolomide versus adjuvant temozolomide: results of a single-institution, retrospective, matched-pair analysisActa Neurochir (Wien)2013155342943523254891
- AdebergSBostelTHarrabiSImpact of delays in initiating postoperative chemoradiation while determining the MGMT promoter-methylation statuses of patients with primary glioblastomaBMC Cancer20151555826223282
- KimYSKimSHChoJMGMT gene promoter methylation as a potent prognostic factor in glioblastoma treated with temozolomide-based chemoradiotherapy: a Single-Institution StudyInt J Radiat Oncol Biol Phys201284366166722414280
- Lechapt-ZalcmanELevalletGDugueAEO(6)-Methylguanine-DNA methyl-transferase (MGMT) promoter methylation and low MGMT-encoded protein expression as prognostic markers in glioblastoma patients treated with biodegradable carmustine wafer implants after initial surgery followed by radiotherapy with concomitant and adjuvant temozolomideCancer2012118184545455422359215
- ReifenbergerGHentschelBFelsbergJPredictive impact of MGMT promoter methylation in glioblastoma of the elderlyInt J Cancer2011131613421350
- FelsbergJThonNEigenbrodSPromoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomasInt J Cancer2011129365967021425258
- BalanaCCarratoCRamirezJLTumour and serum MGMT promoter methylation and protein expression in glioblastoma patientsClin Transl Oncol201113967768521865140
- LakomyRSanaJHankeovaSMiR-195, miR-196b, miR-181c, miR-21 expression levels and O-6-methylguanine-DNA methyltransferase methylation status are associated with clinical outcome in glioblastoma patientsCancer Sci2011102122186219021895872
- SonodaYYokosawaMSaitoRO(6)-Methylguanine DNA methyl-transferase determined by promoter hypermethylation and immunohistochemical expression is correlated with progression-free survival in patients with glioblastomaInt J Clin Oncol201015435235820232102
- MinnitiGSalvatiMArcellaACorrelation between O6-methyl-guanine-DNA methyltransferase and survival in elderly patients with glioblastoma treated with radio-therapy plus concomitant and adjuvant temozolomideJ Neurooncol2011102231131620686820
- ThonNEigenbrodSGrasbon-FrodlEMPredominant influence of MGMT methylation in non-resectable glioblastoma after radiotherapy plus temozolomideJ Neurol Neurosurg Psychiatry201182444144620861061
- WellerMFelsbergJHartmannCMolecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma NetworkJ Clin Oncol200927345743575019805672
- WemmertSBettscheiderMAltSp15 promoter methylation—a novel prognostic marker in glioblastoma patientsInt J Oncol20093461743174819424593
- HegiMEDiserensACGodardSClinical trial substantiates the predictive value of O-6- methylguanine -DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomideClin Cancer Res20041061871187415041700
- ZawlikIVaccarellaSKitaDMittelbronnMFranceschiSOhgakiHPromoter methylation and polymorphisms of the MGMT gene in glioblastomas: a population-based studyNeuroepidemiology2009321212918997474
- ParkCKParkSHLeeSHMethylation status of the MGMT gene promoter fails to predict the clinical outcome of glioblastoma patients treated with ACNU plus cisplatinNeuropathology200929444344919170894
- SonodaYKumabeTWatanabeMLong-term survivors of glioblastoma: clinical features and molecular analysisActa Neurochir2009151111349135819730774
- MetellusPCoulibalyBNanniIPrognostic impact of O6-methylguanine-DNA methyltransferase silencing in patients with recurrent glioblastoma multiforme who undergo surgery and carmustine wafer implantation: a prospective patient cohortCancer2009115204783479419637364
- CaoVTJungTYJungSThe correlation and prognostic significance of MGMT promoter methylation and MGMT protein in glioblastomasNeurosurgery200965586687519834398
- IaccarinoCOrlandiERuggeriFPrognostic value of MGMT promoter status in non-resectable glioblastoma after adjuvant therapyClin Neurol Neurosurg20151321825723791
- BaraultLAmatuABleekerFEDigital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancerAnn Oncol20152691994199926113646
- ShenDLiuTLinQMGMT promoter methylation correlates with an overall survival benefit in Chinese high-grade glioblastoma patients treated with radiotherapy and alkylating agent-based chemotherapy: a single-institution studyPLoS One201499e10755825211033
- MelguizoCPradosJGonzálezBMGMT promoter methylation status and MGMT and CD133 immunohistochemical expression as prognostic markers in glioblastoma patients treated with temozolomide plus radiotherapyJ Transl Med201210125023245659
- KanemotoMShirahataMNakaumaAPrognostic prediction of glioblastoma by quantitative assessment of the methylation status of the entire MGMT promoter regionBMC Cancer201414164125175833
- ChristiansAHartmannCBennerAPrognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastomaPLoS One201273e3344922428052
- CombsSERiekenSWickWPrognostic significance of IDH-1 and MGMT in patients with glioblastoma: one step forward and one step back?Radiat Oncol20116111521910919
- YangSHLeeKSYangHJO(6)-Methylguanine-DNA-methyltransferase promoter methylation assessment by microdissection-assisted methylation-specific PCR and high resolution melting analysis in patients with glioblastomasJ Neurooncol2012106224325021792731
- EllingsonBMCloughesyTFPopeWBAnatomic localization of O6-methylguanine DNA methyl- transferase (MGMT) promoter methylated and unmethylated tumors: a radio-graphic study in 358 de novo human glioblastomasNeuroimage201259290891622001163
- ParkCKKimJYimSYUsefulness of MS-MLPA for detection of MGMT promoter methylation in the evaluation of pseudoprogression in glioblastoma patientsNeuro Oncol201113219520221075779
- CostaBMCaeiroCGuimaraesIPrognostic value of MGMT promoter methylation in glioblastoma patients treated with temozolomide- based chemoradiation: a Portuguese multicentre studyOncol Rep20102361655166220428822
- BrandesAAFranceschiETosoniAO(6)-Methylguanine DNA-methyltransferase methylation status can change between first surgery for newly diagnosed glioblastoma and second surgery for recurrence: clinical implicationsNeuro Oncol201012328328820167816
- MorandiLFranceschiEde BiaseDPromoter methylation analysis of O6-methylguanine-DNA methyltransferase in glioblastoma: detection by locked nucleic acid based quantitative PCR using an imprinted gene (SNURF) as a referenceBMC Cancer20101014820167086
- RiveraALPelloskiCEGilbertMRMGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastomaNeuro Oncol201012211612120150378
- ZunarelliEBigianiNSartoriGMigaldiMSgambatoAMaioranaAINI1 immuno-histochemical expression in glioblastoma: correlation with MGMT gene promoter methylation status and patient survivalPathology2011431172321240060
- Karayan-TaponLQuillienVGuilhotJPrognostic value of O6-methyl-guanine-DNA methyl -transferase status in glioblastoma patients, assessed by five different methodsJ Neurooncol201097331132219841865
- BrandesAAFranceschiETosoniATemozolomide concomitant and adjuvant to radiotherapy in elderly patients with glioblastoma: correlation with MGMT promoter methylation statusCancer2009115153512351819514084
- SmithKAAshbyLSGonzalezLFProspective trial of gross-total resection with Gliadel wafers followed by early postoperative Gamma Knife radiosurgery and conformal fractionated radiotherapy as the initial treatment for patients with radiographically suspected, newly diagnosed glioblastoma multiformeJ Neurosurg2008109suppl10611719123896
- DerSimonianRLairdNMeta-analysis in clinical trialsControl Clin Trials1986731771883802833
- QuillienVLavenuASansonMOutcome-based determination of optimal pyrosequencing assay for MGMT methylation detection in glioblastoma patientsJ Neurooncol2014116348749624420923
- CitronMDeckerRChenSO6-Methylguanine-DNA methyltransferase in human normal and tumor tissue from brain, lung, and ovaryCancer Res19915116413141341868433
- SilberJRMuellerBAEwersTGBergerMSComparison of O6-methyl-guanine-DNA methyltransferase activity in brain tumors and adjacent normal brainCancer Res19935314341634208324751
- ThonNKrethSKrethFWPersonalized treatment strategies in glioblastoma: MGMT promoter methylation statusOnco Targets Ther201361363137224109190
- WickWPlattenMMeisnerCTemozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trialLancet Oncol201213770771522578793
- MalmströmAGrønbergBHMarosiCTemozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trialLancet Oncol201213991692622877848
- HegiMEDiserensACGorliaTMGMT gene silencing and benefit from temozolomide in glioblastomaN Engl J Med200535210997100315758010
- GersonetSLMGMT: its role in cancer aetiology and cancer therapeuticsNat Rev Cancer20044429630715057289
- ZhaoHWangSSongCZhaYLiLThe prognostic value of MGMT promoter status by pyrosequencing assay for glioblastoma patients’ survival: a meta-analysisWorld J of Surg Oncol20161426127733166
- ChakravartiAErkkinenMGNestlerUTemozolomide-mediated radiation enhancement in glioblastoma: a report on underlying mechanismsClin Cancer Res200612154738474616899625
- FianoVTrevisanMTrevisanEMGMT promoter methylation in plasma of glioma patients receiving temozolomideJ Neurooncol2014117234735724519517
- McDonaldKLRapkinsRWOlivierJThe T genotype of the MGMT C[T (rs16906252) enhancer single-nucleotide polymorphism (SNP) is associated with promoter methylation and longer survival in glioblastoma patientsEur J Cancer201349236036822975219
- ChenCHuangRMacLeanARecurrent high-grade glioma treated with bevacizumab: prognostic value of MGMT methylation, EGFR status and pretreatment MRI in determining response and survivalJ Neurooncol2013115226727623974656
- OchsenbeinAFSchubertADVassellaEMarianiLQuantitative analysis of O6-methylguanine DNA methyltransferase (MGMT) promoter methylation in patients with low-grade gliomasJ Neurooncol2011103234335120857319
- StuppRHegiMEGorliaTCilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071-22072 study): a multicentre, randomised, open-label, phase 3 trialLancet Oncol201415101100110825163906
- GlasMHappoldCRiegerJLong-term survival of patients with glioblastoma treated with radiotherapy and lomustine plus temozolo-mideJ Clin Oncol20092781257126119188676
- PradosMDChangSMButowskiNPhase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcomaJ Clin Oncol200927457958419075262
- DunnJBaborieAAlamFExtent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapyBr J Cancer2009101112413119536096
- MuratAMigliavaccaEGorliaTStem cell-related ‘‘self-renewal’’ signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastomaJ Clin Oncol200826183015302418565887
- YangHWeiDYangKTangWLuoYZhangJThe prognosis of MGMT promoter methylation in glioblastoma patients of different race:a meta-analysisNeurochem Res2014391211124258018
- AbhinavKAquilinaKGbejuadeHLaMHopkinsKIyerVA pilot study of glioblastoma multiforme in elderly patients: treatments, O-6-methylguanine-DNA methyltransferase (MGMT) methylation status and survivalClin Neurol Neurosurg201311581375137823333005
- SkiriuteDVaitkienePSaferisVMGMT, GATA6, CD81, DR4, and CASP8 gene promoter methylation in glioblastomaBMC Cancer20121221822672670
- CarrilloJALaiANghiemphuPLRelationship between tumor enhancement, edema, IDH1 mutational status, MGMT promoter methylation, and survival in glioblastomaAJNR Am J Neuroradiol20123371349135522322613
- OhkaFNatsumeAMotomuraKWakabayashi The global DNA methylation surrogate LINE-1 methylation is correlated with MGMT promoter methylation and is a better prognostic factor for gliomaPLoS One201168e2333221829728
- WickWHartmannCEngelCNOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomideJ Clin Oncol200927355874588019901110
- Van den BentMJDubbinkHJSansonMMGMT promoter methylation is prognostic but not predictive for outcome to adjuvant PCV chemotherapy in anaplastic oligodendroglial tumors: a report from EORTC Brain Tumor Group Study 26951J Clin Oncol200927355881588619901104
- StuppRHegiMEMasonWPEffects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trialLancet Oncol200910545946619269895
- SchaichMKestelLPfirrmannMMDR1 (ABCB1) gene single nucleotide polymorphism predicts outcome of temozolomide treatment in glioblastoma patientsAnn Oncol200920117518118687982
- GerstnerERYipSWangDLLouisDNIafrateAJBatchelorTTMgmt methylation is a prognostic biomarker in elderly patients with newly diagnosed glioblastomaNeurology200973181509151019884580
- EoliMMenghiFBruzzoneMGMethylation of O6- methylguanine DNA methyltransferase and loss of heterozygosity on 19q and/or 17p are overlapping features of secondary glioblastomas with prolonged survivalClin Cancer Res20071392606261317473190
- WatanabeTKatayamaYKomineCO6-methylguanine-DNA methyltransferase methylation and TP53 mutation in malignant astro-cytomas and their relationships with clinical courseInt J Cancer2005113458158715455376
- ThonNThorsteinsdottirJEigenbrodSOutcome in unresectable glioblastoma:MGMT promoter methylation makes the differenceJ Neurol2017264235035827921166
- GerstnerERDudaDGdi TomasoEVEGF inhibitors in the treatment of cerebral edema in patients with brain cancerNat Rev Clin Oncol20096422923619333229
- ChengWLiMCaiJA prognostic and chromosomal instability marker, refines the predictive value of MGMT promoter methylationJ Neurooncol2015122230331225557107
