Abstract
Background
Numerous studies have shown that miRNA levels are closely related to the survival time of patients with colon, rectal, or colorectal cancer (CRC). However, the outcomes of different investigations have been inconsistent. Accordingly, a meta-analysis was conducted to study associations among the three types of cancers.
Materials and methods
Studies published in English that estimated the expression levels of miRNAs with survival curves in CRC were identified until May 20, 2017 by online searches in PubMed, Embase, Web of Science, and the Cochrane Library by two independent authors. Pooled HRs with 95% CIs were used to estimate the correlation between miRNA expression and overall survival.
Results
A total of 63 relevant articles regarding 13 different miRNAs, with 10,254 patients were ultimately included. CRC patients with high expression of blood miR141 (HR 2.52, 95% CI 1.68–3.77), tissue miR21 (HR 1.31, 95% CI 1.12–1.53), miR181a (HR 1.52, 95% CI 1.26–1.83), or miR224 (HR 2.12, 95% CI 1.04–4.34), or low expression of tissue miR126 (HR 1.55, 95% CI 1.24–1.93) had significantly poor overall survival (P<0.05).
Conclusion
In general, blood miR141 and tissue miR21, miR181a, miR224, and miR126 had significant prognostic value. Among these, blood miR141 and tissue miR224 were strong biomarkers of prognosis for CRC.
Introduction
Numerous researchers have studied the associations between miRNA expression and the survival outcomes of colorectal cancer (CRC) patients.Citation1–Citation258 CRC has a 10% cancer incidence and mortality worldwide,Citation259 and thus, it is one of the most serious diseases threatening human health. Despite great success in the treatment of CRC, the prognosis of CRC patients is still poor. Therefore, it is fundamental for the diagnosis, treatment, and prognosis of CRC patients to understand its emphasized molecular origin.Citation260 Despite a comprehensive study about the mechanisms of CRC, there are still some challenges that require recognizing prognostic biomarkers with minimal invasion and sensitivity. Accordingly, it is of vital significance to improve the survival rate of CRC patients, utilizing rapid and reliable tumor-prognosis biomarkers.
miRNAs, small noncoding RNA gene products of approximately 22 nucleotides, are found in various types of organisms. They account for 2%–5% of the entire genome, number about 1,000, and regulate the expression of ≥20% of human genes.Citation261 In addition, they play crucial roles in regulating the translation and degradation of mRNAs via base pairing to partially complementary sites, predominantly in the 3′-untranslated areas of mRNAs.Citation262–Citation264
In the study of CRC, a large number of articles have covered the fact that miRNAs are closely related to the survival time of patients.Citation1–Citation258 There were relatively small samples in these papers, and the present work aims to estimate the most accurate prognostic value between miRNA level and survival outcome of CRC patients, better to comprehend the miRNAs with prognostic pertinence that are potential candidates for clinical verification in the future.
Materials and methods
Search strategy
We used four online databases – PubMed, Embase, Web of Science, and the Cochrane Library – to find pertinent literature published until May 20, 2017. The combination term “miR and colorectal cancer” was employed for the literature search. Two authors (S Gao and ZY Zhao) independently performed this comprehensive online search.
Inclusion criteria
Articles qualified if they satisfied the following criteria: patients with colon/rectal cancer or CRC; miRNA levels in tissue, plasma, or serum and survival results were measured; at least one survival curve was measured of overall survival (OS), cause-specific survival (CSS), disease-free survival (DFS), recurrence-free survival (RFS), progression-free survival (PFS), and metastasis-free survival (MFS), with or without HRs/95% CIs; and full text published in English.
Exclusion criteria
Exclusion criteria were experimental studies, reviews, or letters without primary data and retracted papers; frequency of research evaluating prognostic value of miRNAs in tissue of four or less. Only the most comprehensive study was included for this meta-analysis if more than one paper had been published in the same research group.
Quality assessment
SG and ZY Zhao identified all qualifying studies analyzing the prognostic value of miRNAs in CRC, and YZ reevaluated uncertain data.
Study selection
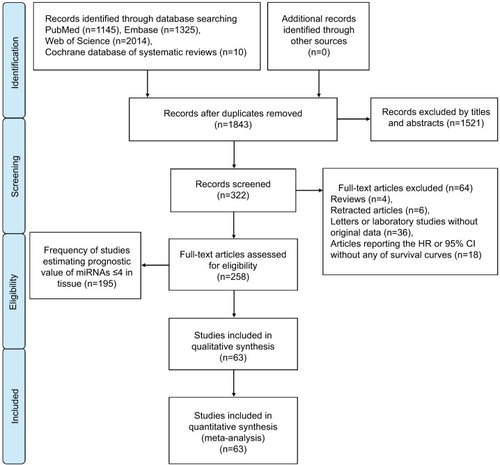
A flow diagram of the study selection process is presented in . Our study found 1,843 articles for consideration within this meta-analysis, and 322 articles suitable for assessment of prognostic miRNA signatures in CRC and full-text papers were acquired by evaluating titles and abstracts. On elaborate review of research methodologies, 64 investigations were excluded, the details of which are shown in . On the basis of the exclusion criteria, 63 studies were finally included in this meta-analysis.
Study frequency
The frequency of studies estimating the prognostic value of miRNAs in CRC are shown in (blood) and 2 (tissue), including miRNA name, number of studies estimating prognostic value, and references.
Table 1 Frequency of studies estimating prognostic value of blood miRNA expression in colorectal cancer
Study characteristics
Literature with Kaplan–Meier survival curves for CRC are detailed in . If data were not provided visually and merely as curves, they were extracted from the curves, and estimated HRs with 95% CIs were subsequently calculated using the method of Tierney et alCitation265 with Engauge Digitizer version 4.1 software. In addition, if outcomes of both univariate and multiple covariates were covered, only the latter was chosen, because of adjustment for confounders.
Table 2 Frequency of studies estimating prognostic value of tissue-miRNA expression in colorectal cancer
Table 3 Characteristics of studies included on colorectal cancer
Statistical analyses
All analyses were performed utilizing Stata version 13.0 (StataCorp, College Station, TX, USA). Merged HRs were regarded as significant at the P<0.05 level if 95% CIs did not contain the value 1. Effect values for HRs were regarded as large if ≥2. HRs for OS were regarded as the prime reference standard if OS P-values were inconsistent with other survival outcomes with respect to the associated miRNA level. All analyses employed random-effect models instead of fixed-effect models, because there existed differences among the studies, including tissue detected (frozen or formalin-fixed, paraffin-embedded), blood (plasma or serum), tumor stage (I–IV), cutoff values, and miRNA-analysis methods. Publication bias was measured by Begg’s funnel plot, and a two-tailed P-value <0.05 was regarded as significant. The trim-and-fill method was performed if publication bias occurred. Sensitivity analysis was employed to weigh how powerful merged HRs were after a single study had been removed. An individual study was suspected of having excess of influence if the point estimation was outside the 95% CI after removal from the analysis.
Results
Meta-analysis
An overview of HRs appraised from comprehensive analysis of all the miRNAs is given in . Thirteen miRNAs were involved in this meta-analysis: miR21, miR92a, miR106a, miR125b, miR126, miR141, miR143, miR145, miR181a, miR200b, miR203, miR224, and miR429. Results of survival analyses of these miRNAs are given in –.
Table 4 Meta-analysis results for miRNA expression in colorectal cancer
Figure 2 Pooled analyses of OS or DFS in association with high blood miR21-, miR92a-, miR141-, miR200b-, and miR203-expression levels.
Note: Weights are from random-effect analysis.
Abbreviations: OS, overall survival; DFS, disease-free survival.
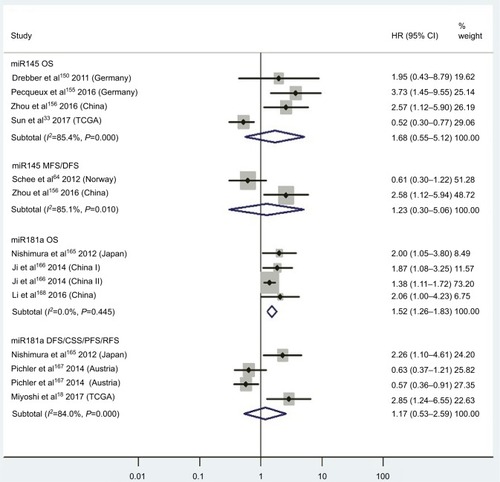
Figure 3 (A) Forest plots of pooled analyses of OS or OS (multiple-covariate analysis) in association with high tissue miR21-expression levels; (B) Begg’s funnel plot of publication bias for pooled analysis of OS in association with high tissue miR21-expression levels; (C) sensitivity analysis of pooled analysis of OS in association with high tissue miR21-expression levels; (D) forest plots of pooled analyses of DFS or RFS/CSS/MFS/PFS in association with high tissue miR21-expression levels. Weights are from random-effects analysis in A and D. aMultiple-covariate analysis; bunivariate analysis.
Abbreviations: OS, overall survival; DFS, disease-free survival; RFS, recurrence-free survival; CSS, cause-specific survival; MFS, metastasis-free survival; PFS, progression-free survival.
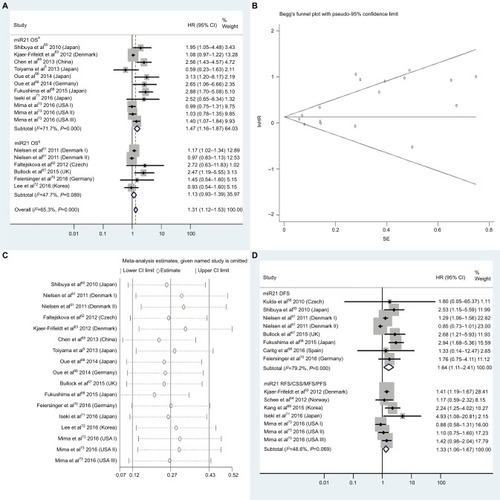
Figure 4 (A) Funnel plot of pooled analysis adjusted with the trim-and-fill method of OS in association with high tissue miR21-expression levels. Circles, included studies; diamonds, presumed missing studies. (B) Forest plot of pooled analysis adjusted with the trim-and-fill method of OS in association with high tissue miR21-expression levels. (C) Sensitivity analysis of pooled analysis adjusted with the trim-and-fill method of OS in association with high tissue miR21-expression levels. Weights are from random-effects analysis. aMultiple-covariate analysis; bunivariate analysis.
Abbreviation: OS, overall survival.
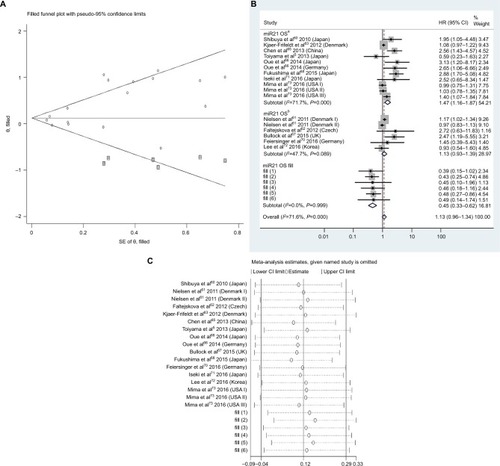
Figure 5 Pooled analyses of OS or DFS/MFS in association with high tissue miR106a- and miR125b-expression levels. Weights are from random-effects analysis.
Abbreviations: OS, overall survival; DFS, disease-free survival; MFS, metastasis-free survival; TCGA, the Cancer Genome Atlas.
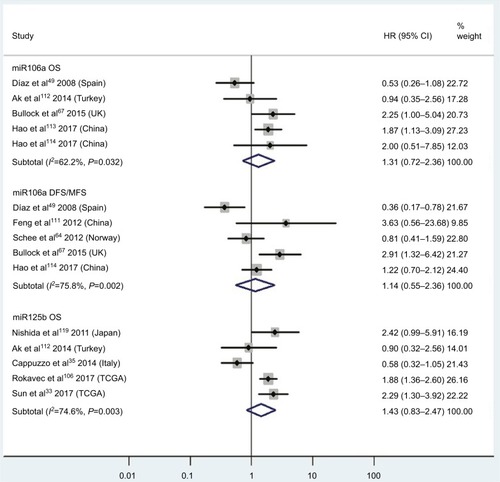
Figure 6 Pooled analyses of OS, PFS/RFS/CSS, or DFS/CSS/PFS in association with low tissue miR126- and miR143-expression levels. Weights are from random-effects analysis.
Abbreviations: OS, overall survival; DCCG, Dutch Colorectal Cancer Group; PFS, progression-free survival; RFS, recurrence-free survival; CSS, cause-specific survival; DFS, disease-free survival.
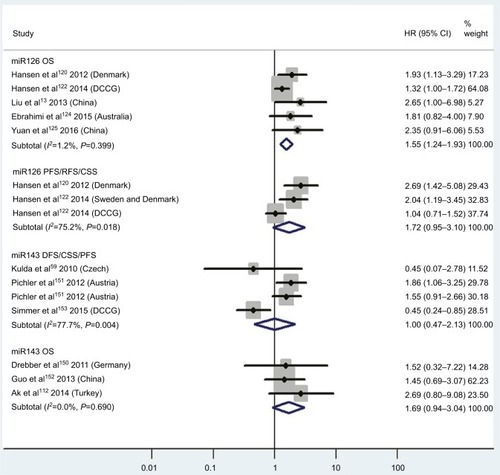
Figure 7 Pooled analyses of OS, MFS/DFS or DFS/CSS/PFS/RFS in association with high tissue miR145-expression levels or low tissue miR181a-expression levels. Weights are from random-effects analysis.
Abbreviations: OS, overall survival; TCGA, the Cancer Genome Atlas; MFS, metastasis-free survival; DFS, disease-free survival; CSS, cause-specific survival; PFS, progression-free survival; RFS, recurrence-free survival.
CRC patients with high blood miR141, high tissue miR181a and miR224, or low tissue miR126 expression have significantly shorter OS
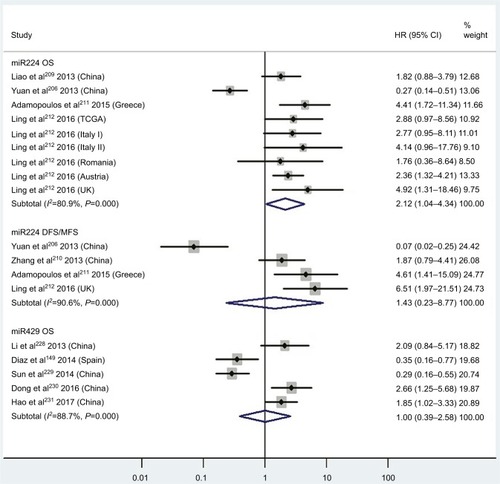
Two studiesCitation14,Citation19 focused on associations between high blood miR141 levels and OS, indicating that CRC patients with high blood miR141 levels had significantly shorter OS than those with low miR141 expression (HR 2.52, 95% CI 1.68–3.77, P<0.01; ). Five papersCitation120,Citation122–Citation125 stressed connections between low tissue miR126 levels and OS, suggesting that CRC patients with low expression of tissue miR126 levels had significantly poorer OS than those with high miR126 expression (HR 1.55, 95% CI 1.24–1.93, P<0.01; ).
Three articles concentrated on the relationship between high tissue miR181a levels and OS, demonstrating that CRC patients with high miR181a levels had significantly worse OS than those with low miR181a expression (HR 1.52, 95% CI 1.26–1.83, P<0.01; ). Four studies paid attention to correlations between high expression of tissue miR224 levels and OS, showing that CRC patients with high tissue miR224 levels had significantly shorter OS than those with low miR224 expression (HR 2.12, 95% CI 1.04–4.34, P=0.04; ).
There was no significant relationship between high expression levels of blood miR21, miR92a, miR200b, miR203, tissue miR106a, miR125b, or miR429 or low expression levels of tissue miR143 or miR145 and OS
Details are given in and and –.
High tissue miR21 expression forecasts poor OS
Thirteen investigationsCitation5,Citation60–Citation68,Citation70–Citation73 analyzed the connection between high tissue miR21 levels and OS, showing that CRC patients with high tissue miR21 levels had significantly worse OS than those with low miR21 expression (HR 1.31, 95% CI 1.12–1.53, P<0.01; ).
Publication bias
To assess publications showing some degree of bias for OS of CRC patients with high tissue miR21 levels, our study used Begg’s funnel plot (). The P-value was less than 0.01, indicating the presence of publication bias. As such, the trim-and-fill method was performed and the pooled HR recalculated with presumed missing studies to estimate asymmetry in the funnel plot (), indicating no publication bias (P=0.73). The recalculated HR changed significance for OS (HR 1.13, 95% CI 0.96–1.34, P=0.15; ).
Sensitivity analysis
For research on OS of CRC patients with high tissue miR21 levels, the sensitivity analysis did not manifest alterations during outcomes on the basis of the exclusion of any single investigation (), showing that no sole study significantly affected the merged HR or 95% CI. This also proved true for the outcome of OS adjusted with the trim-and-fill method ().
Key findings
We carried out a meta-analysis of 13 miRNAs and OS. Serving as the most investigated miRNA, miR21 (high tissue levels) in CRC showed significantly shorter OS than low tissue miR21 levels (P<0.05). However, there was no significant relationship between high blood miR21 levels and OS (P=0.47). The different detected sample types and relatively small sample capacity of the miR21 blood group (only three studies analyzing the relationship between blood miR21 levels and OS) may have been potential clinical reasons and caused the statistical significance between tissue and blood miR21 levels.
Encouragingly, the HR from analysis of the association between high tissue miR21 levels and OS (multiple-covariate analysis)Citation5,Citation60,Citation63,Citation65,Citation66,Citation68,Citation71,Citation73 was 1.47, which was greater than that reported in any of the 13 articles.Citation5,Citation60–Citation78,Citation70–Citation73 Nevertheless, the significance did not remain in accordance with the forest plot, which was adjusted with the trim-and-fill method because publication bias existed (P<0.01; ). This result indicated that the prognostic value of tissue miR21 was not stable in CRC patients. There were other miRNAs with significant prognostic value in CRC, including blood miR141 and tissue miR21, miR181a, miR224, and miR126 (P<0.05). Among these, blood miR141 and tissue miR224 were powerful prognostic candidates in CRC (HR ≥2).
Discussion
Present situation
Increasing numbers of studies have indicated that diverse miRNAs are connected with survival results in CRC patients.Citation1–Citation258 Nevertheless, no systematic review or meta-analysis has evaluated HRs between miRNA levels and survival outcomes of CRC patients. Therefore, it was of vital significance to launch a meta-analysis to comprehend the relationship between expression levels of miRNAs and prognoses of CRC patients.
Molecular mechanisms for miRNAs researched
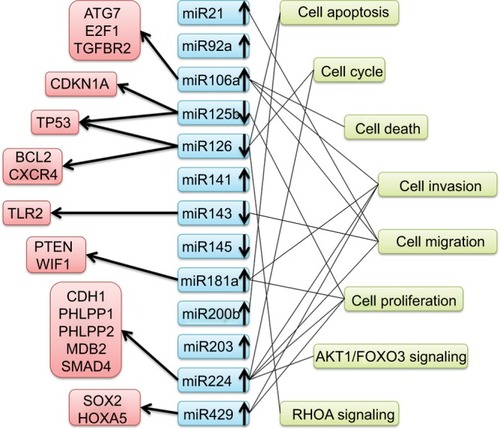
An overview of miRNAs with dysregulated expression and their potential targets and pathways of entry is detailed in . There was noticeable functional overlap and relationships among the miRNAs. Seven miRNAs (miR21, miR106a, miR126, miR143, miR181a, miR224, and miR429) touched upon cell functions, including cell apoptosis, cell cycle, and death. To sum up, these associations may refer to CRC progression.
Figure 9 Summary of microRNAs with altered expression and potential targets and pathways entered in this study.
Abbreviations: ATG7, autophagy related 7; E2F1, E2F transcription factor 1; TGFBR2, transforming growth factor beta receptor 2; CDKN1A, cyclin dependent kinase inhibitor 1A; TP53, tumor protein p53; BCL2, apoptosis regulator; CXCR4, C-X-C motif chemokine receptor 4; TLR2, toll like receptor 2; PTEN, phosphatase and tensin homolog; WIF1, WNT inhibitory factor 1; CDH1, cadherin 1; PHLPP1, PH domain and leucine rich repeat protein phosphatase 1; PHLPP2, PH domain and leucine rich repeat protein phosphatase 2; MBD2, methyl-CpG binding domain protein 2; SMAD4, SMAD family member 4; SOX2, SRY-box 2; HOXA5, homeobox A5; AKT1, AKT serine/threonine kinase 1; FOXO3, forkhead box O3; RHOA, ras homolog family member A.
Other CRC molecular pathways
In addition to miRNAs, there are some other molecular data that can be confounders, related to mortalities, such as the chromosomal instability pathway, the DNA mismatch repair system, and microsatellite instability (MSI). Features of distinctive pathways are different models of genetic instability, succeeding clinical presentations, and features of pathological behavior. A majority of CRC follows the chromosomal instability pathway, features of which are extensive loss of heterozygosis and gross chromosomal abnormalities.Citation266,Citation267 Second, about 15% of CRC is due to the derangement of the DNA mismatch repair system and consequential MSI. The former is in charge of protein production, which identifies and directly repairs mononucleotide mismatches at MS sequences that escape the proofreading system of DNA polymerase. Furthermore, a previous meta-analysis indicated that MSI-high CRC patients had a 40% better OS rate compared with MS-stable CRC patients.Citation268
Molecular pathological epidemiology (MPE)
MPE is a multidisciplinary research field of associations between endogenous and exogenous ingredients, molecular cancer biomarkers, and cancer progression and also a comprehensive interdisciplinary science on the strength of the characteristic principal and continuum theory of diseases.Citation269,Citation270 Other than miRNAs, DNA mutation and methylation and other diagnostics, such as blood tests, also play crucial roles in cancer prognosis and MPE, which deeply investigates environmental exposure, intermediate phenotypes, such as blood biomarkers, and molecular changes in cancer using molecular pathologic analyses. MPE helps precision medicine by providing robust evidence for exposure–outcome associations, such as with drugs.
Strengths
This study has some strengths. Almost all the articles with survival consequences in CRC patients with disparate miR-NAs were searched. Furthermore, the current expression profile of miRNAs is explicitly detailed in and according to miRNAs and types of detected samples (blood or tissue). Papers assessing at least one of the survival curves of OS, CSS, DFS, RFS, PFS, and MFS were eventually included, and papers covering merely HRs or 95% CIs without any of the survival curves were excluded. Meta-analyses were performed on miRNAs investigated five or more times in CRC tissues. Virtually all the studies included had sample sizes ≥30 (except two),Citation70,Citation111 reinforcing the usability and enlarging the feasibility of consequences to CRC patients.
Limitations
Nevertheless, we cannot overemphasize the following limitations. There was much heterogeneity in designs of studies, and most of the outcomes from our meta-analyses contained high heterogeneity (I2≥50%). Statistical assessment of publication bias was suboptimal. There existed differences among the studies, including tissue-detected (frozen or formalin-fixed, paraffin-embedded), blood (plasma or serum), tumor stage (I–IV), cutoff values, and miRNA methods. The present meta-analysis simply included papers published in English, perhaps excluding potential studies published in other languages with respect to miRNA level and prognosis of CRC patients. Papers covering only HRs or 95% CIs without survival curves were excluded, lowering the sample sizes of the papers included. Because of the massive interrelation between papers and data about CRC, we subjectively and selectively included specific studies on the basis of the inclusion and exclusion criteria, bringing about the omission of several possible miRNAs with prognostic value and a relatively small number of included studies. The studies included contained three types of cancers (colon and rectal cancer and CRC), which blurred the division between tumor types. Some blood miRNAs were from cell-free RNA, while others were from exosome isolates. These were considered the same to some degree and may have caused some deviations in the final results.
Implications for prospective clinical and scientific study
It should be mentioned that the current meta-analysis is the first system assessment of the pertinence of miRNA level to the prognosis of CRC patients. This study presents foundations for prospective clinical and scientific study with respect to clinical staff and other health care providers, for whom simultaneous determination of miRNA expression is able greatly to reinforce assessment of life expectancy of CRC patients, thus enabling prompt therapy, and for scientific researchers. Current research progress and trends in connections between miRNAs and prognosis of CRC patients are shown in and . Selectively basic experiments can be conducted using these details (). Conflicting results on the prognosis of miRNAs may be addressed based on the present meta-analysis.
Conclusion
In general, blood miR141 and tissue miR21, miR181a, miR224, and miR126 have significant prognostic value. Among these, blood miR141 and tissue miR224 are strong biomarkers of prognosis in CRC.
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- LiJChenYGuoXInhibition of miR-15b decreases cell migration and metastasis in colorectal cancerTumour Biol20163778765877326743779
- LiJLiuYWangCSerum miRNA expression profile as a prognostic biomarker of stage II/III colorectal adenocarcinomaSci Rep201551292126250939
- MatsumuraTSugimachiKIinumaHExosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancerBr J Cancer2015113227528126057451
- MenéndezPPadillaDVillarejoPPrognostic implications of serum microRNA-21 in colorectal cancerJ Surg Oncol2013108636937323970420
- ToiyamaYTakahashiMHurKSerum miR-21 as a diagnostic and prognostic biomarker in colorectal cancerJ Natl Cancer Inst20131051284985923704278
- MonzoMMartínez-RodenasFMorenoIDifferential MIR-21 expression in plasma from mesenteric versus peripheral veins: an observational study of disease-free survival in surgically resected colon cancer patientsMedicine (Baltimore)2015941e14525569638
- TsukamotoMIinumaHYagiTMatsudaKHashiguchiYCirculating exosomal microRNA-21 as a biomarker in each tumor stage of colorectal cancerOncology201792636037028376502
- KouCHZhouTHanXLZhuangHJQianHXDownregulation of mir-23b in plasma is associated with poor prognosis in patients with colorectal cancerOncol Lett20161264838484428101227
- JinushiTShibayamaYKinoshitaILow expression levels of microRNA-124-5p correlated with poor prognosis in colorectal cancer via targeting of SMC4Cancer Med2014361544155225081869
- WangLGGuJSerum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasisCancer Epidemiol2012361e61e6722018950
- BasatiGRazaviAEPakzadIMalayeriFACirculating levels of the miRNAs, miR-194, and miR-29b, as clinically useful biomarkers for colorectal cancerTumour Biol20163721781178826318304
- SchouJVRossiSJensenBVmiR-345 in metastatic colorectal cancer: a non-invasive biomarker for clinical outcome in non-KRAS mutant patients treated with 3rd line cetuximab and irinotecanPLoS One201496e9988624940606
- LiuGHZhouZGChenRSerum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancerTumour Biol20133442175218123625654
- SunYLiuYCogdellDExamining plasma microRNA markers for colorectal cancer at different stagesOncotarget2016710114341144926863633
- MaoLFengWYuYXuXSerum expression of miRNA-103, a potential diagnostic and prognostic biomarker for colorectal cancerInt J Clin Med2016971421214218
- MaierthalerMBennerAHoffmeisterMPlasma miR-122 and miR-200 family are prognostic markers in colorectal cancerInt J Cancer2017140117618727632639
- KijimaTHazamaSTsunedomiRMicroRNA-6826 and-6875 in plasma are valuable non-invasive biomarkers that predict the efficacy of vaccine treatment against metastatic colorectal cancerOncol Rep2017371233027878288
- MiyoshiJTodenSYoshidaKMiR-139-5p as a novel serum biomarker for recurrence and metastasis in colorectal cancerSci Rep201774339328262692
- ChengHZhangLCogdellDECirculating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosisPLoS One201163e1774521445232
- LvZCFanYSChenHBZhaoDWInvestigation of microRNA-155 as a serum diagnostic and prognostic biomarker for colorectal cancerTumour Biol20153631619162525528214
- YuanDLiKZhuKYanRDangCPlasma miR-183 predicts recurrence and prognosis in patients with colorectal cancerCancer Biol Ther201516226827525629978
- XuCGuLThe diagnostic effect of serum miR-196b as biomarker in colorectal cancerBiomed Rep201761394528123705
- ToiyamaYHurKTanakaKSerum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancerAnn Surg2014259473574323982750
- HurKToiyamaYOkugawaYCirculating microRNA-203 predicts prognosis and metastasis in human colorectal cancerGut201766465466526701878
- ShiSQKeJJWuWQXuQSSerum miRNA-203 expression is associated with chemo-response to standard FOLFOX treatment of patients with colorectal cancerInt J Clin Exp Pathol2017101105116
- PuXXHuangGLGuoHQCirculating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expressionJ Gastroenterol Hepatol201025101674168020880178
- YuJJinLJiangLSerum miR-372 is a diagnostic and prognostic biomarker in patients with early colorectal cancerAnticancer Agents Med Chem201616442443126179262
- HurKToiyamaYSchetterAJIdentification of a metastasis-specific microRNA signature in human colorectal cancerJ Natl Cancer Inst20151073dju49225663689
- ImaokaHToiyamaYFujikawaHCirculating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancerAnn Oncol201627101879188627502702
- LiuCEngCShenJSerum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancerOncotarget2016746762507626027788488
- LiuTPHuangCCYehKTDown-regulation of let-7a-5p predicts lymph node metastasis and prognosis in colorectal cancer: implications for chemotherapySurg Oncol201625442943427262492
- XuMKuangYWangMHanXYangQA microRNA expression signature as a predictor of survival for colon adenocarcinomaNeoplasma2017641566427881005
- SunMSongHWangSIntegrated analysis identifies microRNA-195 as a suppressor of Hippo-YAP pathway in colorectal cancerJ Hematol Oncol20171017928356122
- YangIPTsaiHLMiaoZFDevelopment of a deregulating microRNA panel for the detection of early relapse in postoperative colorectal cancer patientsJ Transl Med201614110827126129
- CappuzzoFSacconiALandiLMicroRNA signature in metastatic colorectal cancer patients treated with anti-EGFR monoclonal antibodiesClin Colorectal Cancer201413137.e445.e424503111
- Molina-PineloSCarneroARiveraFMiR-107 and miR-99a-3p predict chemotherapy response in patients with advanced colorectal cancerBMC Cancer20141465625197016
- SalendoJSpitznerMKramerFIdentification of a microRNA expression signature for chemoradiosensitivity of colorectal cancer cells, involving miRNAs-320a, -224, -132 and let7gRadiother Oncol2013108345145723932154
- KahlertCKluppFBrandKInvasion front-specific expression and prognostic significance of microRNA in colorectal liver metastasesCancer Sci2011102101799180721722265
- SutoTYokoboriTYajimaRMicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulationCarcinogenesis201536333834525503932
- NaganoYToiyamaYOkugawaYMicroRNA-7 is associated with malignant potential and poor prognosis in human colorectal cancerAnticancer Res201636126521652627919977
- ZhuMXuYGeMGuiZYanFRegulation of UHRF1 by microRNA-9 modulates colorectal cancer cell proliferation and apoptosisCancer Sci2015106783383925940709
- NishidaNYamashitaSMimoriKMicroRNA-10b is a prognostic indicator in colorectal cancer and confers resistance to the chemotherapeutic agent 5-fluorouracil in colorectal cancer cellsAnn Surg Oncol20121993065307122322955
- PizziniSBisogninAMandruzzatoSImpact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasisBMC Genomics20131458923987127
- JiangHLiuJChenYMaCLiBHaoTUp-regulation of mir-10b predicate advanced clinicopathological features and liver metastasis in colorectal cancerCancer Med20165102932294127592860
- KontosCKTsiakanikasPAvgerisMPapadopoulosINScorilasAmiR-15a-5p, a novel prognostic biomarker, predicting recurrent colorectal adenocarcinomaMol Diagn Ther201721445346428405803
- XiaoGTangHWeiWLiJJiLGeJAberrant expression of microRNA-15a and microRNA-16 synergistically associates with tumor progression and prognosis in patients with colorectal cancerGastroenterol Res Pract2014201436454925435873
- QianJJiangBLiMChenJFangMPrognostic significance of microRNA-16 expression in human colorectal cancerWorld J Surg201337122944294924045965
- DiamantopoulosMAKontosCKKerimisDPapadopoulosINScorilasAUpregulated miR-16 expression is an independent indicator of relapse and poor overall survival of colorectal adenocarcinoma patientsClin Chem Lab Med201755573774727930363
- DíazRSilvaJGarcíaJMDeregulated expression of miR-106a predicts survival in human colon cancer patientsGenes Chromosomes Cancer200847979480218521848
- MaYZhangPWangFElevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130Nat Commun20123129123250421
- FangLLiHWangLMicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expressionOncotarget20145102974298724912422
- YuGTangJQTianMLPrognostic values of the miR-17-92 cluster and its paralogs in colon cancerJ Surg Oncol2012106323223722065543
- MotoyamaKInoueHTakatsunoYOver- and under-expressed microRNAs in human colorectal cancerInt J Oncol20093441069107519287964
- WuCWDongYJLiangQYMicroRNA-18a attenuates DNA damage repair through suppressing the expression of ataxia telangi-ectasia mutated in colorectal cancerPLoS One201382e5703623437304
- ChenXShiKWangYClinical value of integrated-signature miRNAs in colorectal cancer: miRNA expression profiling analysis and experimental validationOncotarget2015635375443755626462034
- ChengDZhaoSTangHMicroRNA-20a-5p promotes colorectal cancer invasion and metastasis by downregulating Smad4Oncotarget2016729451994521327286257
- ZhangGJLiYZhouHXiaoHXZhouTmiR-20a is an independent prognostic factor in colorectal cancer and is involved in cell metastasisMol Med Rep201410128329124737193
- CaritgONavarroAMorenoIIdentifying high-risk stage II colon cancer patients: a three-microRNA-based score as a prognostic biomarkerClin Colorectal Cancer2016154e175e18227247088
- KuldaVPestaMTopolcanORelevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastasesCancer Genet Cytogenet2010200215416020620599
- ShibuyaHIinumaHShimadaRHoriuchiAWatanabeTClinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancerOncology2010793–431332021412018
- NielsenBSJørgensenSFogJUHigh levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patientsClin Exp Metastasis2011281273821069438
- FaltejskovaPBesseASevcikovaSClinical correlations of miR-21 expression in colorectal cancer patients and effects of its inhibition on DLD1 colon cancer cellsInt J Colorectal Dis201227111401140822476768
- Kjaer-FrifeldtSHansenTFNielsenBSThe prognostic importance of miR-21 in stage II colon cancer: a population-based studyBr J Cancer201210771169117423011541
- ScheeKBoyeKAbrahamsenTWFodstadØFlatmarkKClinical relevance of microRNA miR-21, miR-31, miR-92a, miR-101, miR-106a and miR-145 in colorectal cancerBMC Cancer20121250523121918
- ChenTHChangSWHuangCCThe prognostic significance of APC gene mutation and miR-21 expression in advanced-stage colorectal cancerColorectal Dis201315111367137423773491
- OueNAnamiKSchetterAJHigh miR-21 expression from FFPE tissues is associated with poor survival and response to adjuvant chemotherapy in colon cancerInt J Cancer201413481926193424122631
- BullockMDPickardKMitterRStratifying risk of recurrence in stage II colorectal cancer using deregulated stromal and epithelial microRNAsOncotarget2015697262727925788261
- FukushimaYIinumaHTsukamotoMMatsudaKHashiguchiYClinical significance of microRNA-21 as a biomarker in each Dukes’ stage of colorectal cancerOncol Rep201533257358225421755
- KangWKLeeJKOhSTLeeSHJungCKStromal expression of miR-21 in T3-4a colorectal cancer is an independent predictor of early tumor relapseBMC Gastroenterol201515225609245
- FeiersingerFNolteEWachSMiRNA-21 expression decreases from primary tumors to liver metastases in colorectal carcinomaPLoS One2016112e014858026845148
- IsekiYShibutaniMMaedaKPrognostic significance of microRNA-21 expression in patients with unresectable metastatic colon cancerAnticancer Res201636105145515127798874
- LeeKSNamSKKohJStromal expression of microRNA-21 in advanced colorectal cancer patients with distant metastasesJ Pathol Transl Med201650427027727240857
- MimaKNishiharaRYangJMicroRNA MIR21 (miR-21) and PTGS2 expression in colorectal cancer and patient survivalClin Cancer Res201622153841384826957558
- ZhangGXiaSTianHLiuZZhouTClinical significance of miR-22 expression in patients with colorectal cancerMed Oncol20122953108311222492279
- LiBLiBSunHZhangHThe predicted target gene validation, function, and prognosis studies of miRNA-22 in colorectal cancer tissueTumour Biol2017393 1010428317692257
- ZhouXXuXWangJLinJChenWIdentifying miRNA/mRNA negative regulation pairs in colorectal cancerSci Rep201551299526269151
- WuSBaiLLiZFLiQGXieJJianBClinical significance and prognostic value of microRNA-23b expression level in colon cancerInt J Clin Exp Pathol20169101058710592
- GaoYLiuYDuLDown-regulation of miR-24–3p in colorectal cancer is associated with malignant behaviorMed Oncol201532136225502080
- KerimisDKontosCKChristodoulouSPapadopoulosINScorilasAElevated expression of miR-24-3p is a potentially adverse prognostic factor in colorectal adenocarcinomaClin Biochem201750628529227939727
- LiXYangCWangXZhangJZhangRLiuRThe expression of miR-25 is increased in colorectal cancer and is associated with patient prognosisMed Oncol201431178124293092
- XuJZhaoJZhangRFour microRNAs signature for survival prognosis in colon cancer using TCGA dataSci Rep201663830627974852
- de RobertisMLoiaconoLFusilliCDysregulation of EGFR pathway in EphA2 cell subpopulation significantly associates with poor prognosis in colorectal cancerClin Cancer Res201723115917027401248
- Weissmann-BrennerAKushnirMYanaiGLTumor microRNA-29a expression and the risk of recurrence in stage II colon cancerInt J Oncol20124062097210322426940
- TangWZhuYGaoJMicroRNA-29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E-cadherin via KLF4Br J Cancer2014110245045824281002
- InoueAYamamotoHUemuraMMicroRNA-29b is a novel prognostic marker in colorectal cancerAnn Surg Oncol201522Suppl 3S1410S141825472644
- YangLHYinSYHeRQProspective target genes and pathways of miR-30a-5p in colorectal cancer: an investigation using TCGA and bioinformatics analysisInt J Clin Exp Med201710343734385
- ZhangQTangQQinDRole of microRNA 30a targeting insulin receptor substrate 2 in colorectal tumorigenesisMol Cell Biol2015356988100025582198
- LiaoWTYeYPZhangNJMicroRNA-30b functions as a tumour suppressor in human colorectal cancer by targeting KRAS, PIK3CD and BCL2J Pathol2014232441542724293274
- YanLQiuJYaoJDownregulation of microRNA-30d promotes cell proliferation and invasion by targeting LRH-1 in colorectal carcinomaInt J Mol Med20173961371138028440426
- ManceauGImbeaudSThiébautRHsa-miR-31-3p expression is linked to progression-free survival in patients with KRAS wild-type metastatic colorectal cancer treated with anti-EGFR therapyClin Cancer Res201420123338334724771647
- MlcochovaJFaltejskova-VychytilovaPFerracinMMicroRNA expression profiling identifies miR-31-5p/3p as associated with time to progression in wild-type RAS metastatic colorectal cancer treated with cetuximabOncotarget2015636386953870426497852
- IgarashiHKuriharaHMitsuhashiKAssociation of microRNA-31-5p with clinical efficacy of anti-EGFR therapy in patients with metastatic colorectal cancerAnn Surg Oncol20152282640264825472647
- KissIMlcochovaJBortlicekZEfficacy and toxicity of panitumumab after progression on cetuximab and predictive value of MiR-31-5p in metastatic wild-type KRAS colorectal cancer patientsAnticancer Res20163694955495927630355
- YangMHYuJChenNElevated microRNA-31 expression regulates colorectal cancer progression by repressing its target gene SATB2PLoS One2013812e8535324386467
- ChenTYaoLQShiQMicroRNA-31 contributes to colorectal cancer development by targeting factor inhibiting HIF-1α (FIH-1)Cancer Biol Ther201415551652324521875
- NoshoKIgarashiHNojimaMAssociation of microRNA-31 with BRAF mutation, colorectal cancer survival and serrated pathwayCarcinogenesis201435477678324242331
- WuWYangPFengXThe relationship between and clinical significance of microRNA-32 and phosphatase and tensin homologueGenes Chromosomes Cancer201352121133114024123284
- LiaoWGuCHuangAYaoJSunRMicroRNA-33b inhibits tumor cell growth and is associated with prognosis in colorectal cancer patientsClin Transl Oncol201618544945626329295
- GaoJLiNDongYmiR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorec-tal cancerOncogene201534314142415225362853
- HiyoshiYSchetterAJOkayamaHIncreased microRNA-34b and-34c predominantly expressed in stromal tissues is associated with poor prognosis in human colon cancerPLoS One2015104e012489925894979
- ZhouTZhangGLiuZXiaSTianHOverexpression of miR-92a correlates with tumor metastasis and poor prognosis in patients with colorectal cancerInt J Colorectal Dis2013281192422772712
- KeTWWeiPLYehKTChenWTChengYWMiR-92a promotes cell metastasis of colorectal cancer through PTEN-mediated PI3K/AKT pathwayAnn Surg Oncol20152282649265525515201
- XiaoZGDengZSZhangYDZhangYHuangZCClinical significance of microRNA-93 downregulation in human colon cancerEur J Gastroenterol Hepatol201325329630123354160
- RessALStiegelbauerVWinterEMiR-96-5p influences cellular growth and is associated with poor survival in colorectal cancer patientsMol Carcinog201554111442145025256312
- RaptiSMKontosCKPapadopoulosINScorilasAHigh miR-96 levels in colorectal adenocarcinoma predict poor prognosis, particularly in patients without distant metastasis at the time of initial diagnosisTumour Biol2016379118151182427044381
- RokavecMHorstDHermekingHCellular model of colon cancer progression reveals signatures of mRNAs, miRNA, lncRNAs, and epigenetic modifications associated with metastasisCancer Res20177781854186728130225
- LiWChangJWangSmiRNA-99b-5p suppresses liver metastasis of colorectal cancer by down-regulating mTOROncotarget2015627244482446226259252
- ChenPXiQWangQWeiPDownregulation of microRNA-100 correlates with tumor progression and poor prognosis in colorectal cancerMed Oncol2014311023525216869
- ZhengYBXiaoKXiaoGCMicroRNA-103 promotes tumor growth and metastasis in colorectal cancer by directly targeting LATS2Oncol Lett20161232194220027602163
- YueBSunBLiuCLong non-coding RNA Fer-1-like protein 4 suppresses oncogenesis and exhibits prognostic value by associating with miR-106a-5p in colon cancerCancer Sci2015106101323133226224446
- FengBDongTTWangLLColorectal cancer migration and invasion initiated by microRNA-106aPLoS One201278e4345222912877
- AkSTuncaBTezcanGMicroRNA expression patterns of tumors in early-onset colorectal cancer patientsJ Surg Res2014191111312224746948
- HaoHLiuLZhangDDiagnostic and prognostic value of miR-106a in colorectal cancerOncotarget2017835038504727926519
- HaoHXiaGWangCZhongFLiuLZhangDmiR-106a suppresses tumor cells death in colorectal cancer through targeting ATG7Med Mol Morphol2017502768527981410
- WangYXLangFLiuYXYangCQGaoHJIn situ hybridization analysis of the expression of miR-106b in colonic cancerInt J Clin Exp Pathol20158178679225755775
- ZhangGJLiJSZhouHXiaoHXLiYZhouTMicroRNA-106b promotes colorectal cancer cell migration and invasion by directly targeting DLC1J Exp Clin Cancer Res2015347326223867
- WangMJLiYWangRDownregulation of microRNA-124 is an independent prognostic factor in patients with colorectal cancerInt J Colorectal Dis201328218318922885837
- QiuZGuoWWangQMicroRNA-124 reduces the pen-tose phosphate pathway and proliferation by targeting PRPS1 and RPIA mRNAs in human colorectal cancer cellsGastroenterology201514961587.e111598.e1126248089
- NishidaNYokoboriTMimoriKMicroRNA miR-1205b is a prognostic marker in human colorectal cancerInt J Oncol20113851437144321399871
- HansenTFSørensenFBLindebjergJJakobsenAThe predictive value of microRNA-126 in relation to first line treatment with capecitabine and oxaliplatin in patients with metastatic colorectal cancerBMC Cancer2012128322397399
- HansenTFChristensenRDAndersenRFSørensenFBJohnssonAJakobsenAMicroRNA-126 and epidermal growth factor-like domain 7-an angiogenic couple of importance in metastatic colorectal cancer: results from the Nordic ACT trialBr J Cancer201310951243125123922111
- HansenTFKjær-FrifeldtSMorgenthalerSThe prognostic value of microRNA-126 and microvessel density in patients with stage II colon cancer: results from a population cohortJ Transl Med20141225425199818
- LiuYZhouYFengXLow expression of microRNA-126 is associated with poor prognosis in colorectal cancerGenes Chromosomes Cancer201453435836524532280
- EbrahimiFGopalanVWahabRLuCTSmithRALamAKDeregulation of miR-126 expression in colorectal cancer pathogenesis and its clinical significanceExp Cell Res2015339233334126455548
- YuanWGuoYQLiXYMicroRNA-126 inhibits colon cancer cell proliferation and invasion by targeting the chemokine (C-X-C motif) receptor 4Oncotarget2016737602306024427517626
- TakahashiYIwayaTSawadaGUp-regulation of NEK2 by microRNA-128 methylation is associated with poor prognosis in colorectal cancerAnn Surg Oncol201421120521224046120
- LuWWangJYangGPosttranscriptional regulation of galectin-3 by miR-128 contributes to colorectal cancer progressionOncotarget201789152421525128146425
- ColangeloTFucciAVotinoCMicroRNA-130b promotes tumor development and is associated with poor prognosis in colorectal cancerNeoplasia20131591086109924027433
- ZhengYBLuoHPShiQmiR-132 inhibits colorectal cancer invasion and metastasis via directly targeting ZEB2World J Gastroenterol201420216515652224914372
- MokutaniYUemuraMMunakataKDown-regulation of microRNA-132 is associated with poor prognosis of colorectal cancerAnn Surg Oncol201623Suppl 559960826868958
- WanTMLamCSNgLThe clinicopathological significance of miR-133a in colorectal cancerDis Markers2014201491928325104873
- WangLLDuLTLiJDecreased expression of miR-133a correlates with poor prognosis in colorectal cancer patientsWorld J Gastroenterol20142032113401134625170220
- AkçakayaPEkelundSKolosenkoImiR-185 and miR-133b deregulation is associated with overall survival and metastasis in colorectal cancerInt J Oncol201139231131821573504
- AzizianAEppingIKramerFPrognostic value of microRNAs in preoperative treated rectal cancerInt J Mol Sci201617456827092493
- XieYSongJZongQDecreased expression of miR-134 and its clinical significance in human colorectal cancerHepatogastroenterology20156213961561926897940
- GaedckeJGradeMCampsJThe rectal cancer microRNAome: microRNA expression in rectal cancer and matched normal mucosaClin Cancer Res201218184919413022850566
- ValeriNBraconiCGaspariniPMicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancerCancer Cell201425446948324735923
- KanSFYangJSSunGXSunJJMicroRNA-135b is associated with tumor progression in colorectal cancerInt J Clin Exp Med20169365336538
- ChenDLWangDSWuWJOverexpression of paxillin induced by miR-137 suppression promotes tumor progression and metastasis in colorectal cancerCarcinogenesis201334480381123275153
- SmithARMarquezRTTsaoWCTumor suppressive microRNA-137 negatively regulates Musashi-1 and colorectal cancer progressionOncotarget2015614125581257325940441
- QinYZXieXCLiuHZLaiHQiuHGeLYScreening and preliminary validation of miRNAs with the regulation of hTERT in colorectal cancerOncol Rep20153362728273625845814
- ZhaoLYuHYiSThe tumor suppressor miR-138–5p targets PD-L1 in colorectal cancerOncotarget2016729453704538427248318
- LongLHuangGZhuHGuoYLiuYHuoJDown-regulation of miR-138 promotes colorectal cancer metastasis via directly targeting TWIST2J Transl Med20131127524171926
- LiuXDuanBDongYMicroRNA-139-3p indicates a poor prognosis of colon cancerInt J Clin Exp Pathol20147118046805225550849
- SongMYinYZhangJMiR-139-5p inhibits migration and invasion of colorectal cancer by downregulating AMFR and NOTCH1Protein Cell201451185186125149074
- GuoHHuXGeSQianGZhangJRegulation of RAP1B by miR-139 suppresses human colorectal carcinoma cell proliferationInt J Biochem Cell Biol20124491465147222642900
- ZhaiHFeslerABaYWuSJuJInhibition of colorectal cancer stem cell survival and invasive potential by hsa-miR-140-5p mediated suppression of Smad2 and autophagyOncotarget2015623197351974625980495
- ZhangWZouCPanLMicroRNA-140-5p inhibits the progression of colorectal cancer by targeting VEGFACell Physiol Biochem20153731123113326402430
- DiazTTejeroRMorenoIRole of miR-200 family members in survival of colorectal cancer patients treated with fluoropyrimidinesJ Surg Oncol2014109767668324510588
- DrebberULayMWedemeyerIAltered levels of the onco-microRNA 21 and the tumor-suppressor microRNAs 143 and 145 in advanced rectal cancer indicate successful neoadjuvant chemoradiotherapyInt J Oncol201139240941521567082
- PichlerMWinterEStotzMDown-regulation of KRAS-interacting miRNA-143 predicts poor prognosis but not response to EGFR-targeted agents in colorectal cancerBr J Cancer2012106111826183222549179
- GuoHChenYHuXQianGGeSZhangJThe regulation of Toll-like receptor 2 by miR-143 suppresses the invasion and migration of a subset of human colorectal carcinoma cellsMol Cancer2013127723866094
- SimmerFVenderboschSDijkstraJRMicroRNA-143 is a putative predictive factor for the response to fluoropyrimidine-based chemotherapy in patients with metastatic colorectal cancerOncotarget2015626229962300726392389
- IwayaTYokoboriTNishidaNDownregulation of miR-144 is associated with colorectal cancer progression via activation of mTOR signaling pathwayCarcinogenesis201233122391239722983984
- PecqueuxMLiebetrauIWerftWA comprehensive microRNA expression profile of liver and lung metastases of colorectal cancer with their corresponding host tissue and its prognostic impact on survivalInt J Mol Sci20161710E175527775664
- ZhouPSunLLiuDLiuCSunLLong non-coding RNA lincRNA-ROR promotes the progression of colon cancer and holds prognostic value by associating with miR-145Pathol Oncol Res201622473374027071407
- ChristensenLLTobiasenHHolmAMiRNA-362-3p induces cell cycle arrest through targeting of E2F1, USF2 and PTPN1 and is associated with recurrence of colorectal cancerInt J Cancer20131331677823280316
- TakahashiMCuatrecasasMBalaguerFThe clinical significance of MiR-148a as a predictive biomarker in patients with advanced colorectal cancerPLoS One2012710e4668423056401
- HibinoYSakamotoNNaitoYSignificance of miR-148a in colorectal neoplasia: downregulation of miR-148a contributes to the carcinogenesis and cell invasion of colorectal cancerPathobiology201582523324126389729
- WangFMaYLZhangPSP1 mediates the link between methylation of the tumour suppressor miR-149 and outcome in colorectal cancerJ Pathol20132291122422821729
- XuKLiuXMaoXMicroRNA-149 suppresses colorectal cancer cell migration and invasion by directly targeting forkhead box transcription factor FOXM1Cell Physiol Biochem201535249951525613903
- MaYZhangPWangFmiR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancerGut201261101447145322052060
- ZhangLPickardKJeneiVmiR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistanceCancer Res201373216435644723950211
- KaiYQiangCXinxinPMiaomiaoZKuailuLDecreased miR-154 expression and its clinical significance in human colorectal cancerWorld J Surg Oncol20151319526048406
- NishimuraJHandaRYamamotoHmicroRNA-181a is associated with poor prognosis of colorectal cancerOncol Rep20122862221222623023298
- JiDChenZLiMMicroRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1Mol Cancer2014138624755295
- PichlerMWinterERessALmiR-181a is associated with poor clinical outcome in patients with colorectal cancer treated with EGFR inhibitorJ Clin Pathol201467319820324098024
- LiZWangHXuZSunYHanJExpression and mechanism of microRNA-181A on incidence and survival in late liver metastases of colorectal cancerOncol Rep20163531403140826750720
- BovellLCShanmugamCPutchaBDThe prognostic value of microRNAs varies with patient race/ethnicity and stage of colorectal cancerClin Cancer Res201319143955396523719259
- YamazakiNKogaYTaniguchiHHigh expression of miR-181c as a predictive marker of recurrence in stage II colorectal cancerOncotarget2017846970698328036302
- LiuHDuLWenZUp-regulation of miR-182 expression in colorectal cancer tissues and its prognostic valueInt J Colorectal Dis201328569770323474644
- RaptiSMKontosCKPapadopoulosINScorilasAEnhanced miR-182 transcription is a predictor of poor overall survival in colorectal adenocarcinoma patientsClin Chem Lab Med20145281217122724615484
- WangSYangMHWangXYLinJDingYQIncreased expression of miRNA-182 in colorectal carcinoma: an independent and tissue-specific prognostic factorInt J Clin Exp Pathol2014763498350325031782
- ZhouTZhangGJZhouHXiaoHXLiYOverexpression of microRNA-183 in human colorectal cancer and its clinical significanceEur J Gastroenterol Hepatol201426222923324150523
- WangZSZhongMBianYHMicroRNA-187 inhibits tumor growth and invasion by directly targeting CD276 in colorectal cancerOncotarget2016728442664427627329595
- ZhangFLuoYShaoZMicroRNA-187, a downstream effector of TGFβ pathway, suppresses Smad-mediated epithelial-mesenchymal transition in colorectal cancerCancer Lett2016373220321326820227
- PichlerMStiegelbauerVVychytilova-FaltejskovaPGenome-wide microRNA analysis identifies miR-188-3p as a novel prognostic marker and molecular factor involved in colorectal carcinogenesisClin Cancer Res20172351323133327601590
- QinSZhuYAiFMicroRNA-191 correlates with poor prognosis of colorectal carcinoma and plays multiple roles by targeting tissue inhibitor of metalloprotease 3Neoplasma2014611273424195505
- KaraayvazMPalTSongBPrognostic significance of miR-215 in colon cancerClin Colorectal Cancer201110434034721752725
- ShanBChenPLiSXuLYuHDecreased expression of microRNA-192 correlates with tumor progression and poor prognosis in patients with colorectal cancerInt J Clin Exp Pathol2017101595602
- ZhangPJiDBHanHBShiYFDuCZGuJDownregulation of miR-193a-5p correlates with lymph node metastasis and poor prognosis in colorectal cancerWorld J Gastroenterol20142034122411224825232258
- GuoFLuoYMuYFmiR-193b directly targets STMN1 and inhibits the malignant phenotype in colorectal cancerAm J Cancer Res20166112463247527904764
- ZhaoHJRenLLWangZHMiR-194 deregulation contributes to colorectal carcinogenesis via targeting AKT2 pathwayTheranostics20144121193120825285168
- WangBShenZLGaoZDMiR-194, commonly repressed in colorectal cancer, suppresses tumor growth by regulating the MAP4K4/c-Jun/MDM2 signaling pathwayCell Cycle20151471046105825602366
- GeJChenZLiRLuTXiaoGUpregulation of microRNA-196a and microRNA-196b cooperatively correlate with aggressive progression and unfavorable prognosis in patients with colorectal cancerCancer Cell Int201414112825525411
- BoisenMKDehlendorffCLinnemannDTissue microRNAs as predictors of outcome in patients with metastatic colorectal cancer treated with first line capecitabine and oxaliplatin with or without bevacizumabPLoS One2014910e10943025329796
- WangMWangJKongXMiR-198 represses tumor growth and metastasis in colorectal cancer by targeting fucosyltransferase 8Sci Rep20144614525174450
- WanDHeSXieBAberrant expression of miR-199a-3p and its clinical significance in colorectal cancersMed Oncol201330137823292866
- ShenZLWangBJiangKWDownregulation of miR-199b is associated with distant metastasis in colorectal cancer via activation of SIRT1 and inhibition of CREB/KISS1 signalingOncotarget2016723350923510527145368
- PichlerMRessALWinterEMiR-200a regulates epithelial to mesenchymal transition-related gene expression and determines prognosis in colorectal cancer patientsBr J Cancer201411061614162124504363
- XiYFormentiniAChienMPrognostic values of microRNAs in colorectal cancerBiomark Insights2006211312118079988
- DengBWangBFangJMiRNA-203 suppresses cell proliferation, migration and invasion in colorectal cancer via targeting of EIF5A2Sci Rep201662830127376958
- SümbülATGöğebakanBErgünSmiR-204-5p expression in colorectal cancer: an autophagy-associated geneTumour Biol20143512127131271925209181
- YinYZhangBWangWmiR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorec-tal cancer cells by downregulating RAB22AClin Cancer Res201420236187619925294901
- SunPSunDWangXLiuTMaZDuanLmiR-206 is an independent prognostic factor and inhibits tumor invasion and migration in colorectal cancerCancer Biomark201515439139626406866
- QuADuLYangYHypoxia-inducible MiR-210 is an independent prognostic factor and contributes to metastasis in colorectal cancerPLoS One201493e9095224632577
- SümbülATGöğebakanBBayramSBatmacıCYÖztuzcuSMicroRNA 211 expression is upregulated and associated with poor prognosis in colorectal cancer: a case-control studyTumour Biol201536129703970926152286
- MengXWuJPanCGenetic and epigenetic down-regulation of microRNA-212 promotes colorectal tumor metastasis via dysregulation of MnSODGastroenterology20131452426.e1-e6436.e1-e623583431
- ChenDLWangZQZengZLIdentification of microRNA-214 as a negative regulator of colorectal cancer liver metastasis by way of regulation of fibroblast growth factor receptor 1 expressionHepatology201460259860924616020
- LiSGaoJGuJYuanJHuaDShenLMicroRNA-215 inhibits relapse of colorectal cancer patients following radical surgeryMed Oncol201330254923532818
- WangBShenZLJiangKWMicroRNA-217 functions as a prognosis predictor and inhibits colorectal cancer cell proliferation and invasion via an AEG-1 dependent mechanismBMC Cancer20151543726016795
- ZhangNLuCChenLmiR-217 regulates tumor growth and apoptosis by targeting the MAPK signaling pathway in colorectal cancerOncol Lett20161264589459728105166
- YuHGaoGJiangLDecreased expression of miR-218 is associated with poor prognosis in patients with colorectal cancerInt J Clin Exp Pathol20136122904291124294377
- LiPLZhangXWangLLMicroRNA-218 is a prognostic indicator in colorectal cancer and enhances 5-fluorouracil-induced apoptosis by targeting BIRC5Carcinogenesis201536121484149326442524
- TaoKYangJGuoZPrognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancerAm J Transl Res20146439140125075256
- YuanKXieKFoxJDecreased levels of miR-224 and the passenger strand of miR-221 increase MBD2, suppressing maspin and promoting colorectal tumor growth and metastasis in miceGastroenterology20131454853.e9864.e923770133
- CaiKShenFCuiJHYuYPanHQExpression of miR-221 in colon cancer correlates with prognosisInt J Clin Exp Med2015822794279825932237
- LiZWYangYMDuLTOverexpression of miR-223 correlates with tumor metastasis and poor prognosis in patients with colorectal cancerMed Oncol2014311125625270282
- LiaoWTLiTTWangZGmicroRNA-224 promotes cell proliferation and tumor growth in human colorectal cancer by repressing PHLPP1 and PHLPP2Clin Cancer Res201319174662467223846336
- ZhangGJZhouHXiaoHXLiYZhouTUp-regulation of miR-224 promotes cancer cell proliferation and invasion and predicts relapse of colorectal cancerCancer Cell Int201313110424152489
- AdamopoulosPGKontosCKRaptiSMPapadopoulosINScorilasAmiR-224 overexpression is a strong and independent prognosticator of short-term relapse and poor overall survival in colorectal adenocarcinomaInt J Oncol201546284985925420464
- LingHPickardKIvanCThe clinical and biological significance of miR-224 expression in colorectal cancer metastasisGut201665697798925804630
- HeZYuLLuoSmiR-296 inhibits the metastasis and epithelial-mesenchymal transition of colorectal cancer by targeting S100A4BMC Cancer201717114028209128
- Perez-CarbonellLSinicropeFAAlbertsSRMiR-320e is a novel prognostic biomarker in colorectal cancerBr J Cancer20151131839026035698
- SchepelerTReinertJTOstenfeldMSDiagnostic and prognostic microRNAs in stage II colon cancerCancer Res200868156416642418676867
- WuLHuiHWangLJWangHLiuQFHanSXMicroRNA-326 functions as a tumor suppressor in colorectal cancer by targeting the Nin one binding proteinOncol Rep20153352309231825760058
- SunZZhangZLiuZQiuBLiuKDongGMicroRNA-335 inhibits invasion and metastasis of colorectal cancer by targeting ZEB2Med Oncol201431698224829139
- SunKSuGDengHDongJLeiSLiGRelationship between miRNA-338-3p expression and progression and prognosis of human colorectal carcinomaChin Med J (Engl)2014127101884189024824250
- TakeyamaHYamamotoHYamashitaSDecreased miR-340 expression in bone marrow is associated with liver metastasis of colorectal cancerMol Cancer Ther201413497698524448820
- MaFSongHGuoBMiR-361-5p inhibits colorectal and gastric cancer growth and metastasis by targeting staphylococcal nuclease domain containing-1Oncotarget2015619174041741625965817
- NieJLiuLZhengWMicroRNA-365, down-regulated in colon cancer, inhibits cell cycle progression and promotes apoptosis of colon cancer cells by probably targeting cyclin D1 and Bcl-2Carcinogenesis201233122022522072615
- YamashitaSYamamotoHMimoriKMicroRNA-372 is associated with poor prognosis in colorectal cancerOncology201282420521222456107
- MoZHWuXDLiSFeiBYZhangBExpression and clinical significance of microRNA-376a in colorectal cancerAsian Pac J Cancer Prev201415219523952725422250
- LiHDaiSZhenTClinical and biological significance of miR-378a-3p and miR-378a-5p in colorectal cancerEur J Cancer20145061207122124412052
- ZhangGJZhouHXiaoHXLiYZhouTMiR-378 is an independent prognostic factor and inhibits cell growth and invasion in colorectal cancerBMC Cancer20141410924555885
- ZhengGXQuALYangYMZhangXZhangSCWangCXmiR-422a is an independent prognostic factor and functions as a potential tumor suppressor in colorectal cancerWorld J Gastroenterol201622245589559727350737
- TorresSGarcia-PalmeroIBartoloméRACombined miRNA profiling and proteomics demonstrates that different miRNAs target a common set of proteins to promote colorectal cancer metastasisJ Pathol20172421395128054337
- LiJDuLYangYMiR-429 is an independent prognostic factor in colorectal cancer and exerts its anti-apoptotic function by targeting SOX2Cancer Lett20133291849023111103
- SunYShenSTangHmiR-429 identified by dynamic transcriptome analysis is a new candidate biomarker for colorectal cancer prognosisOMICS2014181546424237355
- DongSJCaiXJLiSJThe clinical significance of miR-429 as a predictive biomarker in colorectal cancer patients receiving 5-fluorouracil treatmentMed Sci Monit2016223352336127654003
- HanYZhaoQZhouJShiRmiR-429 mediates tumor growth and metastasis in colorectal cancerAm J Cancer Res20177221823328337372
- YeYPWuPGuCCmiR-450b-5p induced by oncogenic KRAS is required for colorectal cancer progressionOncotarget2016738613126132427494869
- HataTMokutaniYTakahashiHIdentification of microRNA-487b as a negative regulator of liver metastasis by regulation of KRAS in colorectal cancerInt J Oncol201750248749628000854
- XuXChenRLiZMicroRNA-490-3p inhibits colorectal cancer metastasis by targeting TGFβR1BMC Cancer201515102326714817
- SunHBChenXJiHmiR-494 is an independent prognostic factor and promotes cell migration and invasion in colorectal cancer by directly targeting PTENInt J Oncol20144562486249425270723
- LiLSarverALKhatriRSequential expression of miR-182 and miR-503 cooperatively targets FBXW7, contributing to the malignant transformation of colon adenoma to adenocarcinomaJ Pathol2014234448850125269767
- NoguchiTToiyamaYKitajimaTmiRNA-503 promotes tumor progression and is associated with early recurrence and poor prognosis in human colorectal cancerOncology201690422123126999740
- ZhangYLinCLiaoGMicroRNA-506 suppresses tumor proliferation and metastasis in colon cancer by directly targeting the oncogene EZH2Oncotarget2015632325863260126452129
- ZhouHLinCZhangYmiR-506 enhances the sensitivity of human colorectal cancer cells to oxaliplatin by suppressing MDR1/P-gp expressionCell Prolif2017503e12341
- MaWYuQJiangJmiR-517a is an independent prognostic marker and contributes to cell migration and invasion in human colorectal cancerOncol Lett20161142583258927073521
- YeCYueGShenZmiR-542-3p suppresses colorectal cancer progression through targeting survivinTransl Cancer Res201656817826
- YuanLYuanPYuanHmiR-542-3p inhibits colorectal cancer cell proliferation, migration and invasion by targeting OTUB1Am J Cancer Res20177115917228123857
- ZhouQZhuYWeiXMiR-590-5p inhibits colorectal cancer angiogenesis and metastasis by regulating nuclear factor 90/vascular endothelial growth factor A axisCell Death Dis2016710e241327735951
- OuCSunZLiXMiR-590-5p, a density-sensitive microRNA, inhibits tumorigenesis by targeting YAP1 in colorectal cancerCancer Lett2017399536328433598
- LiuMZhiQWangWZhangQFangTMaQUp-regulation of miR-592 correlates with tumor progression and poor prognosis in patients with colorectal cancerBiomed Pharmacother20156921422025661360
- ShiFLiRGuoPExpression and prognostic value of miR-610 in patients with colorectal cancerBiomed Res201728313211324
- RasmussenMHJensenNFTarpgaardLSHigh expression of microRNA-625-3p is associated with poor response to first-line oxaliplatin based treatment of metastatic colorectal cancerMol Oncol20137363764623506979
- LouXQiXZhangYLongHYangJDecreased expression of microRNA-625 is associated with tumor metastasis and poor prognosis in patientsJ Surg Oncol2013108423023523861214
- ChuDZhengJLiJMicroRNA-630 is a prognostic marker for patients with colorectal cancerTumour Biol201435109787979224981248
- ZhangJFeiBWangQMicroRNA-638 inhibits cell proliferation, invasion and regulates cell cycle by targeting tetraspanin 1 in human colorectal carcinomaOncotarget2014523120831209625301729
- WangXKuangYShenXEvaluation of miR-720 prognostic significance in patients with colorectal cancerTumour Biol201536271972725286763
- ZhangTCaiXLiQHsa-miR-875-5p exerts tumor suppressor function through down-regulation of EGFR in colorectal carcinoma (CRC)Oncotarget2016727422254224027302926
- LiuDRGuanQLGaoMTJiangLKangHXmiR-1260b is a potential prognostic biomarker in colorectal cancerMed Sci Monit2016222417242327399918
- GopalanVPillaiSEbrahimiFRegulation of microRNA-1288 in colorectal cancer: altered expression and its clinicopathologicalMol Carcinog201453Suppl 1E36E4424009195
- JuHQLuYXChenDLRedox regulation of stem-like cells through the CD44v-xCT Axis in colorectal cancer: mechanisms and therapeutic implicationsTheranostics2016681160117527279909
- HuYYiBHeSClinical significance of miR-1826 as a novel prognostic biomarker in colorectal cancerAnticancer Agents Med Chem20161691109111625968874
- YuFYTuYDengYMiR-4500 is epigenetically downregulated in colorectal cancer and functions as a novel tumor suppressor by regulating HMGA2Cancer Biol Ther201617111149115727686621
- ZhaoSSunHJiangWmiR-4775 promotes colorectal cancer invasion and metastasis via the Smad7/TGFβ-mediated epithelial to mesenchymal transitionMol Cancer20171611228095858
- JemalASiegelRXuJWardECancer statistics, 2010CA Cancer J Clin201060527730020610543
- ValeriNCroceCMFabbriMPathogenetic and clinical relevance of microRNAs in colorectal cancerCancer Genomics Proteomics20096419520419656996
- FormanJJLegesse-MillerACollerHAA search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequenceProc Natl Acad Sci U S A200810539148791488418812516
- Lagos-QuintanaMRauhutRLendeckelWTuschlTIdentification of novel genes coding for small expressed RNAsScience2001294554385385811679670
- LauNCLimLPWeinsteinEGBartelDPAn abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegansScience2001294554385886211679671
- LeeRCAmbrosVAn extensive class of small RNAs in Caenorhabditis elegansScience2001294554386286411679672
- TierneyJFStewartLAGhersiDBurdettSSydesMRPractical methods for incorporating summary time-to-event data into meta-analysisTrials200781617555582
- LinJKChangSCYangYCLiAFLoss of heterozygosity and DNA aneuploidy in colorectal adenocarcinomaAnn Surg Oncol20031091086109414597448
- LearyRJLinJCCumminsJIntegrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancersProc Natl Acad Sci U S A200810542162241622918852474
- GuastadisegniCColafranceschiMOttiniLDogliottiEMicrosatellite instability as a marker of prognosis and response to therapy: a meta-analysis of colorectal cancer survival dataEur J Cancer201046152788279820627535
- OginoSChanATFuchsCSGiovannucciEMolecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary fieldGut201160339741121036793
- OginoSNishiharaRvan der WeeleTJThe role of molecular pathological epidemiology in the study of neoplastic and non-neoplastic diseases in the era of precision medicineEpidemiology201627460261126928707