Abstract
Objective
The aim of this study was to investigate the prognostic value of mircoRNA-17 and mircoRNA-17-5P (miR-17/17-5P) in patients with cancer.
Materials and methods
We conducted a comprehensive search on published literature following the guidelines of the meta-analysis of observational studies in epidemiology group for design, implementation, and reporting. The methodological qualities for included studies were assessed using the quality in prognosis studies. The pooled hazard ratios (HRs) with 95% CIs for overall survival (OS) and progression-free survival/recurrence-free survival/disease-free survival (PFS/RFS/DFS) were calculated to appraise the associations between miR-17/17-5P expression and cancer prognosis.
Results
A total of 21 studies involving 2099 subjects were analyzed in evidence synthesis. The results showed that high expression of miR-17 was associated with poor OS (HR=2.14; 95% CI: 1.69–2.71, P<0.001) in patients with cancer, especially in Caucasian (HR=2.23; 95% CI: 1.58–3.14, P<0.001) and digestive tract cancer (HR=1.29, 95% CI: 1.03–1.63, P=0.03), and miR-17 expression was significantly correlated with PFS/RFS in cancer patients (HR=1.69, 95% CI: 1.29–2.22, P<0.001). miR-17-5P overexpression was significantly linked with poor OS in cancer patients (HR=1.66; 95% CI: 1.31–2.09, P=0.00), especially in Asian (HR=1.81; 95% CI: 1.37–2.40, P<0.001), digestive tract cancer (HR=1.80; 95% CI: 1.29–2.50, P<0.001), and serum sample (HR=1.76; 95% CI: 1.29–2.41, P<0.001). miR-17-5P expression was significantly associated with DFS in cancer patients (HR=1.58, 95% CI: 1.07–2.35, P=0.02).
Conclusion
High expression of miR-17 and miR-17-5P are significantly associated with poor survival in patients with cancer. This indicated that miR-17/17-5P may be a novel prognostic indicator in cancer.
Keywords:
Introduction
Cancer is one of the major disease burdens worldwide.Citation1 In 2016, cancer was the second leading cause of death in the United States.Citation2 Cancer has been the leading cause of death with about one-fourth of all deaths in China.Citation3 The burden of disease attributable to the occurrence of cancers was rising.
MicroRNA (miRNA) is a kind of small, non-protein-coding RNA (containing about 22 nucleotides) molecules involving in RNA silencing and post-transcriptional regulation of gene expression through mRNA degradation and translational repression,Citation4,Citation5 which regulate gene expression by binding to the 3′-untranslated region (UTR) of target mRNAs.Citation6
More and more studies had proved that aberrantly expressed miRNAs were involved in different types of cancers, which acted as tumor oncogenes or suppressors and played critical roles in many aspects of cancer carcinogenesis, including cell proliferation, differentiation, angiogenesis, and metastasis.Citation7–Citation10 miRNAs are regarded as biomarkers for cancer prognosis and prediction of treatment response beacuse of the high stability of miRNAs in circulation and formalin-fixed, paraffin-embedded (FFPE) tissue.Citation11–Citation13
MicroRNA-17 (miR-17) family is the major common studied oncogenic miRNA (onco-miRNA) groups.Citation14 miR-17 and miR-17-5P (miR-17/17-5P) belong to miR-17 family, which has been shown to play a key role in the pathogenesis of diverse cancers.Citation15 miR-17 overexpression significantly improve the ability of migration and motility of melanoma cells by suppression of translation of ETV1.Citation16 The miR-17-5p was overexpressed in ovarian cancer cells. It activated AKT by lowering phosphatase and tensin homolog (PTEN) in ovarian cancer cells.Citation17
High levels of the oncogenic miRNA (onco-miR) guide strand were called miR-17-5p. In triple-negative breast cancer, the overexpressed miR-17-5p can inhibit ribosomal translation of cancer suppressor gene mRNAs, such as PTEN or programmed cell death 4 (PDCD4).Citation18 However, expression of miR-17/17-5P in the circulation or tissue have not been predicted or experimentally confirmed, and the overall survival (OS) and progression-free survival/recurrence-free survival/disease-free survival (PFS/RFS/DFS) of the patterns and expression levels of miR-17/17-5P have not been well established.
In this study, a systematic evaluation with the prognostic data of miR-17/17-5P from published studies was first performed. We focused on correlations between the expression level of miR-17/17-5P and prognostic indicator, which can guide clinical decisions.
Materials and methods
The present study was executed in accordance with the guidelines of the Meta-analysis of Observational Studies in Epidemiology (MOOSE)Citation19 group and criteria of Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA).Citation20 The protocol of this meta-analysis has not been published or registered to any databases.
Search strategy
Studies related to miR-17/17-5P and cancers were retrieved from multiple databases: PubMed, EMBASE, Web of Science, Google Scholar, Wanfang (China), and the Chinese National Knowledge Infrastructure (CNKI) database through July 2017. The search items were combinations of “microRNA-17” or “miR-17”, “microRNA-17-5P” or “miR-17-5P” and “neoplasms” or “cancer”. We additionally manually retrieved bibliography of the selected articles to identify further potentially relevant studies that may be missed by online information retrieval.
Study selection and exclusion criteria
Studies were eligible for inclusion if they were: 1) cohort studies that investigated the relationship between miR-17/17-5P and patients with cancer prognosis; 2) provided available data to extrapolate the hazard ratio for survival and corresponding 95% CI; 3) the expression of miR-17/17-5P was measured in cancer tissue or serum; and 4) available in Chinese or English.
Exclusion criteria were: 1) reviews, experimental cell research, non-human research, letters; 2) neither Chinese nor English studies; 3) experiments not conducted on cancer patients; and 4) insufficient data to calculate the HRs and their 95% CIs, or the Kaplan–Meier curve unable to calculate HRs and 95% CI parameters. If a study had overlapping data with other published literature, we selected the study with a larger sample size or the latest published article. Two independent reviewers (FJD and WGL) screened all initially identified articles according to the eligibility for inclusion criteria.
Data extraction
Two reviewers (YY and JZ) independently identified eligibility studies and performed data extraction and quality assessment. Discrepancies were resolved by consensus and by referencing the original reports.
The following data/necessary information was extracted from the eligible studies if they were available: first author, year of publication, original country, histopathological classification and stage, sample size and type, detection method, cutoff value and follow-up, HRs of miR-17/17-5P for OS and/or PFS/RFS/DFS and their 95% CIs. If the study only provided survival data in a Kaplan–Meier curve, the HR and 95% CI were digitized and extracted using the methods designed by Parmar et alCitation21 and Tierney et al.Citation22
Quality assessment
The quality of included studies was assessed according to the Newcastle–Ottawa Scale (NOS).Citation23 The method rates observational studies on a nine-point scale, and a study with a score ≥6 was considered a high-quality study.
The Quality In Prognosis Studies (QUIPS) for specific biases of prognosis was appraised based on the method of Hayden et al.Citation24 Estimation of the potential bias of the items included study participation, study attrition, prognostic factor measurement, outcome measurement, study confounding, statistical analysis, and reporting.
The quality evaluation was processed independently by two authors (FQL and WGL), and in case of any inconsistency, the final decision was reached with consensus.
Data synthesis and statistical analysis
Data were analyzed by using RRevMan (Version 5.3 for Windows, Cochrane Collaboration, Oxford, UK) and STATA (Version 13.1MP, StataCorp, College Station, TX, USA). HRs and corresponding 95% CIs were utilized to assess the relationship strength between miR-17/17-5P expression and prognosis. Inter-study heterogeneity was evaluated using the Cochran’s Q test and Higgin’s I2. Pheterogeneity<0.1 and I2> 50% represents high heterogeneity. The random-effects model (DerSimonian and Laird method)Citation25 was used in a pooled analysis. Afterward, meta-regression was utilized to explore sources of heterogeneity.Citation26 Otherwise, a fixed-effects model (Mantel–Haenszel method)Citation27 was used. The subgroup analyses were performed by ethnicity (Asian, Caucasian) and cancer subtypes (pathological type), if a cancer type was studied in less than 2 individual studies, it was classified as the “other cancers” group. Assessing the impact of included studies on the pooled results, one-way sensitivity analyses were performed to compensate for statistical heterogeneity.
Begg’s test (rank correlation test)Citation28 and Egger’s test (weighted linear regression test)Citation29 are used to assess the extent of publication bias. A two-tailed value of P<0.05 was considered statistically significant.
Results
Study identification
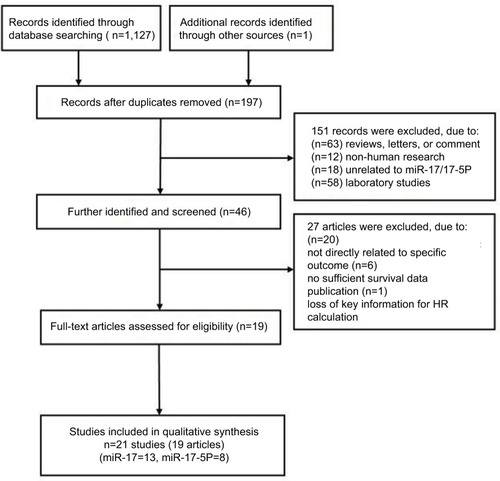
The search process returned 1127 relevant citations and the titles were screened based on the search strategy (). According to the exclusion criteria, the abstracts of 197 studies were reviewed. The titles and abstracts were screened by 2 of the authors. Full texts of potentially eligible articles were retrieved for further identification, 19 eligible articles were screened out. The reference lists of eligible articles were screened for potential publications. Finally, a total of 19 articles (21 studies)Citation30–Citation48 including 11 articles (13 studies)Citation30–Citation40 for miR-17 and 8Citation41–Citation48 for miR-17-5P were considered in a pooled analysis. One of the articlesCitation39 performed three cohort studies in different populations.
Baseline characteristics of included studies
The main characteristics of eligible studies are presented in . The studies were published from 2008 to 2017 and included 2099 patients from China, Brazil, America, Nor-way, Japan, Spain, and Italy. The patients were classified as Asian or Caucasian based on ethnic background. The types of cancer included breast cancer, osteosarcoma, lymphoma, esophageal squamous cell carcinoma (ESCC), oral cancer, glioma, myeloma, colon cancer, non-small cell lung cancer (NSCLC), gastric cancer, colorectal cancer, hepatocellular carcinoma (HCC), endometrial serous adenocarcinoma (ESC) and malignant mesothelioma (MM). The method of miR-17/17-5P detection was quantitative real-time polymerase chain reaction (qRT-PCR) in 21 studies. miR-17/17-5P expression levels were measured in tissue or serum. The cutoff values of miR-17/17-5P were mostly normal or median.
Table 1 Clinicopathological characteristics of eligible studies
Qualitative assessment
The results based on QUIPS are summarized in . The estimated items include the participation, attrition, measurement of prognostic factor, confounding measurement and account, outcome measurement, and analysis and reporting 6 bias domains and the risk of bias shown in and . According to the NOS (), 84% (16/19) of these articles were high-quality (quality score ≥ 6).
Table 2 Quality assessment of included studies based on the Quality in Prognosis Studies (QUIPS)
Test of heterogeneity
The results of heterogeneity tests are shown in . There was no significant heterogeneity among studies for the miR-17(I2 =23%, P=0.21) and miR-17-5P (I2 =0%, P=0.53) in association between miR-17/17-5P expression and the risk of tumorigenesis. Therefore, the fixed effects were applied to calculate the pooled HR for miR-17/17-5P.
Table 3 Main results of pooled HRs in the meta-analysis
Meta-analysis findings
We applied a fixed-effects model to evaluate the pooled HR value (95% CI).
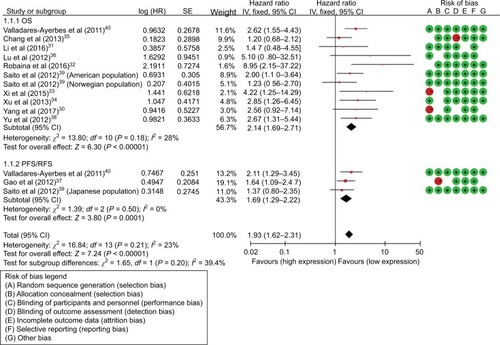
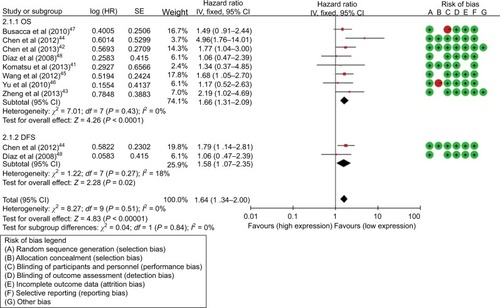
As for the miR-17, HRs of OS were provided by 11 studies, and a significant relation was observed between high miR-17 expression levels and poor OS (HR=2.14; 95% CI: 1.69–2.71, P<0.001). HRs of disease progression were provided by 3 studies and the miR-17 expression level was significantly correlated with PRS/RFS in cancers (HR=1.69; 95% CI: 1.29–2.22, P<0.001) (, ). Subgroup analysis was conducted by ethnicity and miR-17 expression was significantly associated with OS in Asian (HR=2.06; 95% CI: 1.49–2.87, P<0.001) and Caucasian (HR=2.23; 95% CI: 1.58–3.14, P<0.001) (). Additionally, subgroup analysis was carried out according to cancer subtype and the results indicated that a high expression level of miR-17 significantly predicted poor OS in digestive tract cancer (DTC) (HR=2.10; 95% CI: 1.54–2.87, P<0.001) and other cancer types (HR=2.20; 95% CI: 1.53–3.16, P<0.001) ().
As for the miR-17-5P, HRs of OS were provided by 8 studies, and a significant association was observed between high miR-17-5P expression level and poor OS (HR=1.66; 95% CI: 1.31–2.09, P<0.001) (, ). HRs for disease progression were provided by 2 studies, and miR-17-5P expression was significantly correlated with DFS in cancers (HR=1.58; 95% CI: 1.07–2.35, P=0.02) (, ). Subgroups analyses were carried out by ethnicity and miR-17-5P expression level was significantly correlated with OS in Asian (HR=1.81; 95% CI: 1.37–2.40, P<0.001), but not in Caucasian (HR=1.36; 95% CI: 1.89–2.07, P=0.15). Subgroups analyses were also carried out based on cancer subtypes and miR-17-5P expression was significantly correlated with OS in DTC (HR=1.80; 95% CI: 1.29–2.50, P<0.001) and other cancer types (HR=1.53; 95% CI: 1.10–2.13, P<0.001). Furthermore, subgroups analyses were performed according to sample types and the result indicated that a high expression level of miR-17 significantly predicted poor OS in tissue sample (HR=1.53; 95% CI: 1.08–2.18, P=0.02) and serum sample (HR=1.76; 95% CI: 1.29–2.41, P<0.001) ().
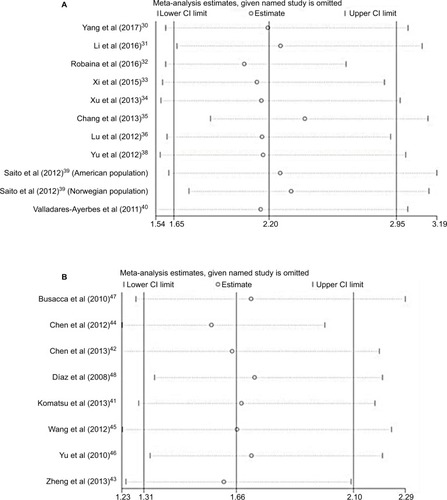
Sensitivity analyses
Sensitivity analysis was performed to assess the contribution of each study to the pooled estimate by omitting individual data sets and recalculating the pooled HR estimates for the remaining studies. The stable pooled HR was found not to be dominantly influenced by any individual study ().
Publication bias
Begg’s and Egger’s tests were used to assess the publication bias. No significant publication bias was found in the included studies (). Meanwhile, the shape of the funnel plots indicated no obvious publication bias (Figures S1 and S2).
Table 4 Publication bias of miR-17/17-5P for Begg’sCitation28 test and Egger’sCitation29 test
Discussion
Emerging studies have demonstrated that miRNAs can act as oncogenes and tumor suppressor genes and play a crucial role in several processes, including cell proliferation, apoptosis, differentiation and metastasis.Citation49,Citation50 Therefore, exploring the profiles of miRNAs and corresponding target genes involved in tumorigenesis may promote the understanding of underlying mechanisms of tumor formation and provide valuable insights for the treatment and early diagnosis of such diseases.Citation51,Citation52 Results from published studies have continually indicated that the miRNAs which are present in the circulation can be promising diagnostic and prognostic biomarkers for tumors.Citation53,Citation54
In this study, increased expression of miR-17 and miR-17-5P were found to be good predictors of poor survival in patients with a variety of cancers. The combined HRs of OS were 2.14 (95% CI: 1.69–2.71) for miR-17 and 1.69 (95% CI: 1.31–2.09) for miR-17-5P, indicating that elevated miR-17/17-5P levels are closely linked with the prognosis of patients with different types of malignant tumors. This is particularly true for miR-17 in the Caucasian, whose pooled HR of OS was 2.23 (95% CI: 1.58–3.14), and this result further demonstrated the predictive value of miR-17. Subgroup analyses also indicated a closer relationship between rising miR-17 levels and poor survival in the Caucasian subgroup. Among 19 studies reporting on OS in 12 cancer types, 10 were DTC. Due to the limited number of included studies for each cancer type, we carried out a stratified analysis of DTC. The result also revealed that elevated miR-17/17-5P yielded worse OS in DTC (HR=2.10; 95% CI: 1.54–2.87 for miR-17; HR=1.80; 95% CI: 1.29–2.50 for miR-17-5P). Meanwhile, further research is needed to explore whether pathological types of specific cancers affect the prognostic role of miR-17/17-5P.
Because the eligible studies used various indices to assess cancer progression, such as DFS, PRS, and RFS, we combined these prognostic indicators to evaluate the prognostic value of miR-17/17-5P. The results indicated an intimate association between high miR-17 expression levels and PFS/RFS (pooled HR=1.69; 95% CI: 1.29–2.22) and high miR-17-5P expression levels and DFS (pooled HR=1.58; 95% CI: 1.07–2.35). Therefore, high miR-17/17-5P expression levels may be a promising negative prognostic factor in DTC. In addition, subgroup analyses were performed according to sample types. However, further study is needed to confirm the role of miR-17/17-5P in predicting the prognosis of different types of cancer.
Evidence has suggested a controversial role of the miR-17 in tumorigenesis, tumor suppressive,Citation55,Citation56 or oncogenic,Citation57,Citation58 which suggested a tumor/tissue-specific role of miR-17. The miR-17-5p functionally modulates sensitivity to radiation in vitro in esophageal adenocarcinoma cells and alters expression of predicted miR-17-5p target genes, such as C6orf120.Citation59 In vivo, miR-17-5p is decreased significantly, while target gene expression is increased significantly in pretreatment tumor biopsies from patients who have a poor response to neoadjuvant chemoradiation therapy.Citation60 It indicates that miR-17-5p can be regarded as a predictive marker of response to neoadjuvant chemoradiation therapy and novel therapeutic target to enhance efficacy of esophageal adenocarcinoma in chemoradiation therapy.Citation61 Meanwhile, compared to normal tissue, miR-17/17-5P have been found at higher levels in colorectal cancerous tissue, although with varied expression of each individual component.Citation38,Citation62 A similar result was observed by Wang et al,Citation45 who demonstrated that the circulating miR-17-5p expression level might be a molecular marker for gastric cancer.
As a comprehensive review of prognostic studies, one limitation is the heterogeneous nature of the identified studies. The reviewed studies varied in study quality, study populations, and statistical analyses. The influencing factors of heterogeneity included in analysis model, definitions of measurement, pathologic classification, laboratory techniques, and treatments received.Citation63 In some situations, inadequate data from some types of cancer and subgroup analyses in HRs may be the origin of heterogeneity.Citation64 The miRNAs can induce widely variable effects depending on tumor biology, microenvironment, and a multitude of other interconnected factors. If the number of studies is very small, it may be impossible to estimate the between-studies variance (τ2) with any precision.Citation65
To our knowledge, there is no evidence-based meta-analysis to evaluate the prognostic values of miR-17/17-5P. Therefore, we pooled the available evidence from all relevant studies to assess the prognostic values of miR-17/17-5P. Although our meta-analysis is robust, several limitations deserved focus as follows. First, not all the included studies provide multivariate adjusted HRs and corresponding 95CI%. In this case, the data was extracted from Kaplan–Meier survival curves, which can result in several tiny errors on calculated HRs with the 95% CIs. Second, although we find no evidence of publication bias, included studies were mostly in English, which may generate publication bias. Third, the cutoff values (median, mean, etc.) were applied to evaluate the different miR-17/17-5P expression, but the actual values may be discrepant due to different algorithms and result in some heterogeneity. Diversity of anatomical locations can be considered as another potential reason for heterogeneity. Fourth, given the limited availability of included studies, for DFS/PFS/RFS, the included studies were not stratified. Finally, the influence of adjuvant therapies on the prognostic effect of malignant tumor was not assessed in our study due to scant data. In spite of these limitations, the study about the relationship between miR-17/17-5P and prognosis of cancers is certainly warranted. In summary, the high expression of miR-17/17-5P was significantly associated with poor survival in patients with cancer. In addition, overexpression of miR-17/17-5P was associated with ethnicity, cancer subtypes, and sample types. These findings suggested that miR-17/17-5P might be a novel prognostic indicator of cancer. Given its limitations, the results of the present analysis should be interpreted with caution. Further clinical investigations are needed to determine the association between miR-17/17-5P and cancer prognosis.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 81701536) and Zhengzhou University Joint First Affiliated Hospital Cultivation Fund (No. 2016-BSTDJJ-15).
Supplementary materials
Figure S1 (A) Begg’s funnel plot of publication bias on the relationship between miR-17 expression and OS.Citation20 (B) Egger’s funnel plot of publication bias on the relationship between miR-17 expression and OS.Citation21
Abbreviations: HR, hazard ratio; miR, microRNA; SND, standard normal deviate.
Figure S2 (A) Begg’s funnel plot of publication bias on the relationship between miR-17-5P expression and OS.Citation20 (B) Egger’s funnel plot of publication bias on the relationship between miR-17-5P expression and OS.Citation21
Abbreviations: HR, hazard ratio; miR, microRNA; SND, standard normal deviate.
Table S1 Quality assessment of included studies based on the Newcastle–Ottawa Scale for assessing the quality of cohort studies
References
- YangFYuanLXuLZhuYGaoHZhenLFanLmiR-17 as a diagnostic biomarker regulates cell proliferation in breast cancerOnco Targets Ther20171054355028203087
- LiSGaoYWangYSerum microRNA-17 functions as a prognostic biomarker in osteosarcomaOncol Lett20161264905491028105199
- RobainaMCFaccionRSMazzoccoliLmiR-17-92 cluster components analysis in Burkitt lymphoma: overexpression of miR-17 is associated with poor prognosisAnn Hematol201695688189127044389
- XiYLiJZhangPUpregulation of miRNA-17 and miRNA-19 is associated with unfavorable prognosis in patients with T-cell lymphoblastic lymphomaExp Mol Pathol201599229730226231295
- XuXLJiangYHFengJGSuDChenPCMaoWMMicroRNA-17, microRNA-18a, and microRNA-19a are prognostic indicators in esophageal squamous cell carcinomaAnn Thorac Surg20149731037104524360091
- ChangCCYangYJLiYJCorrigendum to “MicroRNA-17/20a functions to inhibit cell migration and can be used a prognostic marker in oral squamous cell carcinoma” [Oral Oncol. 2013;49(9):923–931]Oral Oncol20177220220328711305
- LuSWangSGengSMaSLiangZJiaoBIncreased expression of microRNA-17 predicts poor prognosis in human gliomaJ Biomed Biotechnol2012201297076123226946
- GaoXZhangRQuXMiR-15a, miR-16-1 and miR-17-92 cluster expression are linked to poor prognosis in multiple myelomaLeukemia Res201236121505150922959509
- YuGTangJQTianMLPrognostic values of the miR-17-92 cluster and its paralogs in colon cancerJ Surg Oncol2012106323223722065543
- SaitoMSchetterAJMollerupSThe association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohortsClin Cancer Res20111771875188221350005
- Valladares-AyerbesMBlancoMHazMPrognostic impact of disseminated tumor cells and microRNA-17-92 cluster deregulation in gastrointestinal cancerInt J Oncol20113951253126421743960
- KomatsuSIchikawaDTsujiuraMPrognostic impact of circulating miR-21 in the plasma of patients with gastric carcinomaAnticancer Res201333127127623267156
- ChenQSiQXiaoSPrognostic significance of serum miR-17-5p in lung cancerMed Oncol201330135323263848
- ZhengJDongPGaoSWangNYuFHigh expression of serum miR-17-5p associated with poor prognosis in patients with hepatocellular carcinomaHepatogastroenterology20136012354955223108086
- ChenLJiangMYuanWTangHmiR-17-5p as a novel prognostic marker for hepatocellular carcinomaJ Invest Surg201225315616122583011
- WangMGuHWangSCirculating miR-17-5p and miR-20a: molecular markers for gastric cancerMol Med Rep2012561514152022406928
- YuJOhuchidaKMizumotoKFujitaHNakataKTanakaMMicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasionCancer Biol Ther201010874875720703102
- BusaccaSGermanoSDe CeccoLMicroRNA signature of malignant mesothelioma with potential diagnostic and prognostic implicationsAm J Respir Cell Mol Biol201042331231919502386
- DíazRSilvaJGarcíaJMDeregulated expression of miR-106a predicts survival in human colon cancer patientsGenes Chromosomes Cancer200847979480218521848
- BeggCBMazumdarMOperating characteristics of a rank correlation test for publication biasBiometrics1994504108811017786990
- EggerMDavey SmithGSchneiderMMinderCBias in meta-analysis detected by a simple, graphical testBMJ199731571096296349310563
Disclosure
The authors report no conflicts of interest in this work.
References
- SiegelRLMillerKDJemalACancer statistics, 2017CA Cancer J Clin201767173028055103
- SiegelRLMillerKDJemalACancer statistics, 2016CA Cancer J Clin2016661726742998
- ChenWZhengRBaadePDCancer statistics in China, 2015CA Cancer J Clin201666211513226808342
- SunKLaiECAdult-specific functions of animal microRNAsNat Rev Genet201314853554823817310
- LuJGetzGMiskaEAMicroRNA expression profiles classify human cancersNature2005435704383483815944708
- AmbrosVThe functions of animal microRNAsNature2004431700635035515372042
- WinbanksCEOoiJYNguyenSSMcMullenJRBernardoBCMicroRNAs differentially regulated in cardiac and skeletal muscle in health and disease: Potential drug targets?Clin Exp Pharmacol Physiol201441972773725115402
- VorvisCKoutsioumpaMIliopoulosDDevelopments in miRNA gene signaling pathways in pancreatic cancerFuture Oncol20161291135115026984178
- LinCWLinPYYangPCNoncoding RNAs in tumor epithelial-to-mesenchymal transitionStem Cells Int20162016273270526989421
- WalterRFHVollrechtCWernerRMicroRNAs are differentially regulated between MDM2-positive and negative malignant pleural mesotheliomaOncotarget2016714187131872126918730
- TreeceALDuncanDLTangWGastric adenocarcinoma microRNA profiles in fixed tissue and in plasma reveal cancer-associated and Epstein–Barr virus-related expression patternsLab Invest201696666126950485
- GomesBCSantosBRueffJRodriguesASMethods for studying microRNA expression and their targets in formalin-fixed, paraffin-embedded (FFPE) breast cancer tissuesMethods Mol Biol2016139518920526910075
- WenJYeFHeXDevelopment and validation of a prognostic nomogram based on the log odds of positive lymph nodes (LODDS) for breast cancerOncotarget2016715210462105326992235
- KogaYYamazakiNYamamotoYFecal miR-106a is a useful marker for colorectal cancer patients with false-negative results in immunochemical fecal occult blood testCancer Epidemiol Biomarkers Prev201322101844185223950216
- TangYZhangYChenYXiangYShenCLiYThe role of miR-19b in the inhibition of endothelial cell apoptosis and its relationship with coronary artery diseaseSci Rep201551513226459935
- FangYXuCFuYMicroRNA-17-5p induces drug resistance and invasion of ovarian carcinoma cells by targeting PTEN signalingJ Biol Res (Thessalon)2015221211025984506
- CohenRGreenbergENemlichYSchachterJMarkelGmiR-17 regulates melanoma cell motility by inhibiting the translation of ETV1Oncotarget2015622190061901626158900
- JinYYAndradeJWickstromENon-specific blocking of miR-17-5p guide strand in triple negative breast cancer cells by amplifying passenger strand activityPloS one20151012e014257426629823
- StroupDFBerlinJAMortonSCMeta-analysis of observational studies in epidemiology: A proposal for reporting. Meta-analysis of observational studies in epidemiology (MOOSE) groupJAMA2000283152008201210789670
- MoherDLiberatiATetzlaffJAltmanDGPRISMA GroupPreferred reporting items for systematic reviews and meta-analyses: the PRISMA statementAnn Intern Med2009151426426919622511
- ParmarMKTorriVStewartLExtracting summary statistics to perform meta-analyses of the published literature for survival endpointsStat Med19981724281528349921604
- TierneyJFStewartLAGhersiDBurdettSSydesMRPractical methods for incorporating summary time-to-event data into meta-analysisTrials2007811617555582
- WellsGSheaBO’ConnellDThe Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-AnalysesOttawa, ON, CanadaThe Ottawa Hospital Research Institute2011
- HaydenJAvan der WindtDACartwrightJLCôtéPBombardierCAssessing bias in studies of prognostic factorsAnn Intern Med2013158428028623420236
- DerSimonianRLairdNMeta-analysis in clinical trialsControl Clin Trials1986731771883802833
- ThompsonSGHigginsJPHow should meta-regression analyses be undertaken and interpreted?Stat Med200221111559157312111920
- MantelNHaenszelWStatistical aspects of the analysis of data from retrospective studies of diseaseJ Natl Cancer Inst195922471974813655060
- BeggCBMazumdarMOperating characteristics of a rank correlation test for publication biasBiometrics1994504108811017786990
- EggerMDavey SmithGSchneiderMMinderCBias in meta-analysis detected by a simple, graphical testBMJ199731571096296349310563
- YangFYuanLXuLmiR-17 as a diagnostic biomarker regulates cell proliferation in breast cancerOnco Targets Ther20171054355028203087
- LiSGaoYWangYSerum microRNA-17 functions as a prognostic biomarker in osteosarcomaOncol Lett20161264905491028105199
- RobainaMCFaccionRSMazzoccoliLmiR-17-92 cluster components analysis in Burkitt lymphoma: overexpression of miR-17 is associated with poor prognosisAnn Hematol201695688189127044389
- XiYLiJZhangPUpregulation of miRNA-17 and miRNA-19 is associated with unfavorable prognosis in patients with T-cell lymphoblastic lymphomaExp Mol Pathol201599229730226231295
- XuXLJiangYHFengJGSuDChenPCMaoWMMicroRNA-17, microRNA-18a, and microRNA-19a are prognostic indicators in esophageal squamous cell carcinomaAnn Thorac Surg20149731037104524360091
- ChangCCYangYJLiYJCorrigendum to “MicroRNA-17/20a functions to inhibit cell migration and can be used a prognostic marker in oral squamous cell carcinoma” [Oral Oncol499201392393123602254
- Oral Oncol20177220220328711305
- LuSWangSGengSMaSLiangZJiaoBIncreased expression of microRNA-17 predicts poor prognosis in human gliomaJ Biomed Biotechnol2012201297076123226946
- GaoXZhangRQuXmiR-15a, miR-16-1 and miR-17-92 cluster expression are linked to poor prognosis in multiple myelomaLeukemia Res201236121505150922959509
- YuGTangJQTianMLPrognostic values of the miR-17-92 cluster and its paralogs in colon cancerJ Surg Oncol2012106323223722065543
- SaitoMSchetterAJMollerupSThe association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohortsClin Cancer Res20111771875188221350005
- Valladares-AyerbesMBlancoMHazMPrognostic impact of disseminated tumor cells and microRNA-17-92 cluster deregulation in gastrointestinal cancerInt J Oncol20113951253126421743960
- KomatsuSIchikawaDTsujiuraMPrognostic impact of circulating miR-21 in the plasma of patients with gastric carcinomaAnticancer Res201333127127623267156
- ChenQSiQXiaoSPrognostic significance of serum miR-17-5p in lung cancerMed Oncol201330135323263848
- ZhengJDongPGaoSWangNYuFHigh expression of serum miR-17-5p associated with poor prognosis in patients with hepatocellular carcinomaHepatogastroenterology20136012354955223108086
- ChenLJiangMYuanWTangHmiR-17-5p as a novel prognostic marker for hepatocellular carcinomaJ Invest Surg201225315616122583011
- WangMGuHWangSCirculating miR-17-5p and miR-20a: Molecular markers for gastric cancerMol Med Rep2012561514152022406928
- YuJOhuchidaKMizumotoKFujitaHNakataKTanakaMMicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasionCancer Biol Ther201010874875720703102
- BusaccaSGermanoSDe CeccoLMicroRNA signature of malignant mesothelioma with potential diagnostic and prognostic implicationsAm J Respir Cell Mol Biol201042331231919502386
- DíazRSilvaJGarcíaJMDeregulated expression of miR-106a predicts survival in human colon cancer patientsGenes Chromosomes Cancer200847979480218521848
- ChoWCMicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapyInt J Biochem Cell Biol20104281273128120026422
- ChoWCOncomiRs: the discovery and progress of microRNAs in cancersMol Cancer200766017894887
- BrowneGTaipaleenmäkiHSteinGSSteinJLLianJBMicroRNAs in the control of metastatic bone diseaseTrends Endocrinol Metab201425632032724811921
- BryantRJPawlowskiTCattoJWFChanges in circulating microRNA levels associated with prostate cancerBr J Cancer2012106476877422240788
- JoosseSAMüllerVSteinbachBPantelKSchwarzenbachHCirculating cell-free cancer-testis MAGE-A RNA, BORIS RNA, let-7b and miR-202 in the blood of patients with breast cancer and benign breast diseasesBr J Cancer2014111590991724983365
- ZampetakiAKiechlSDrozdovIPlasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetesCirc Res2010107681081720651284
- YuZWangCWangMA Cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferationJ Cell Biol2008182350951718695042
- ZhengZFSuHFZouYPengZWuSXExpression profiles of microRNAs in radioresistant esophageal cell lineZhonghua Yi Xue Za Zhi2011919639642 In Chinese21600139
- DewsMHomayouniAYuDAugmentation of tumor angiogenesis by a Myc-activated microRNA clusterNat Genet20063891060106516878133
- HossainAKuoMTSaundersGFmir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNAMol Cell Biol200626218191820116940181
- ZhangWLinJWangPSunJmiR-17-5p down-regulation contributes to erlotinib resistance in non-small cell lung cancer cellsJ Drug Target201725212513127633093
- YangXDuWWLiHBoth mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasionNucleic Acids Res201341219688970423990326
- Lynam-LennonNHeaveySSommervilleGMicroRNA-17 is downregulated in esophageal adenocarcinoma cancer stem-like cells and promotes a radioresistant phenotypeOncotarget2016871140011413
- DiosdadoBWielMAVDDrosteJSTSmiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progressionBr J Cancer2009101470771419672269
- Cuyun CarterGBarrettAMKayeJALiepaAMWinfreeKBJohnWJA comprehensive review of nongenetic prognostic and predictive factors influencing the heterogeneity of outcomes in advanced non-small-cell lung cancerCancer Manag Res2014643744925364274
- BarkerEVCervigneNKReisPPmicroRNA evaluation of unknown primary lesions in the head and neckMol Cancer2009812720028561
- JamaliZAsl AminabadiNAttaranRPournagiazarFGhertasi OskoueiSAhmadpourFMicroRNAs as prognostic molecular signatures in human head and neck squamous cell carcinoma: a systematic review and meta-analysisOral Oncol201551432133125677760