Abstract
Objective
To explore the relationship between single-nucleotide polymorphisms (SNPs) in one of the long noncoding RNA (lncRNA), cancer susceptibility candidate 8 (CASC8) gene and the risk of cancer.
Materials and methods
A meta-analysis was conducted to summarize the relationship between common SNPs (rs10505477 and rs7837328) in the lncRNA CASC8 gene and the risk of cancer. The relevant references were retrieved from several authoritative databases. Rigorous inclusion and exclusion criteria were adopted to ensure the credibility of the results. The fixed effects or random effects model was used to calculate the OR and 95% CI. We tested for publication bias.
Results
Fifteen articles containing 20 datasets (24,504 cases and 22,969 controls) were finally included in the meta-analysis. Compared to the individuals carrying the rs10505477 TT genotype, those with the TC or CC genotype had a decreased risk of cancer (TC vs TT: OR 0.876, 95% CI 0.832–0.923, P<0.001; CC vs TT: OR 0.748, 95% CI 0.703–0.795, P<0.001). Allele C of rs10505477 might be a protective factor for decreasing susceptibility to cancer (OR 0.866, 95% CI 0.840–0.893, P<0.001). As for rs7837328, the GA and AA genotypes were associated with increased risks of cancer as compared to the GG genotype (ORs 1.209 and 1.336; 95% CIs 1.127–1.298 and 1.202–1.484, respectively); its A allele could significantly increase the risk of cancer compared with the G allele (OR 1.169, 95% CI 1.114–1.227, P<0.001).
Conclusion
The rs10505477 and rs7837328 polymorphisms might be associated with risk of cancer.
Background
Long non-coding RNA (lncRNA) is a noncoding RNA class with a length of more than 200 nucleotides (nt). Previous studies have shown that lncRNA plays an important role in many cellular processes such as cell cycle, apoptosis, epigenetics, and regulation of gene expression; in addition, lncRNA has become a research hotspot in the genetic and molecular epidemiology fields.Citation1,Citation2 In recent years, it has been found that lncRNA expression or functional abnormalities are closely related to the occurrence of human diseases, including several serious diseases that harm human health such as cancer and degenerative neurological diseases; specific manifestations were presented in the abnormal expression of lncRNA in sequential and spatial structures. A previous study showed that lncRNAs could be regarded as noninvasive tumor biomarkers in urologic malignancies, and their alterations could promote tumor development in prostate, bladder, and kidney cancers.Citation3 In fact, it is worth deeply studying not only tumors of the urinary system but also the relationship between lncRNA and other tumors. The focus of this study is to explore the association between the genetic variants located in one lncRNA (cancer susceptibility candidate 8 [CASC8]) and the risks of various types of cancer.
The CASC8 gene is located in the 8q24 – a non-protein-coding region including plenty of genetic loci. Recent studies have revealed that the lncRNAs originating from the human 8q24 locus play important roles in MYC regulation, which is known to be a key contributor to the development of many human tumors.Citation4 The expression of CASC8 is suggested to be significantly correlated with increased cancer susceptibility. Moreover, this study found that the MYC enhancer region physically interacts with the active regulatory region of the CASC8 promoter, suggesting that long-range interaction of the MYC enhancer with the CASC8 promoter regulates CASC8 expression. Finally, Kim et al demonstrated that CARLo-5 has a function in cell-cycle regulation and tumor development.Citation5 Genome-wide association studies (GWAS) and several case–control studies have proved that some particular variants in CASC8 had correlation with carcinomas such as breast cancer, colorectal cancer (CRC), prostate cancer (pCa), upper gastrointestinal cancer, lung cancer, and gastric cancer.Citation6–Citation10 The single-nucleotide polymorphism (SNP) of rs10505477 – located in the intron of the lncRNA of CASC8 gene – had an intimate correlation with CRC susceptibility,Citation11–Citation13 the risk of lung cancer, the prognosis for gastric cancer,Citation14,Citation6 and so on. Rs7837328 – another polymorphism in CASC8 – was associated with pCa and CRC susceptibility.Citation7,Citation15 Although the above studies have reported the association between polymorphisms in the CASC8 gene and risks of cancer, the results were not consistent. Thus, the effect of polymorphisms in the CASC8 gene on cancer is still unclear. Therefore, we conducted an updated meta-analysis on all available studies to assess the overall cancer risk with rs10505477 and rs7837328.
Materials and methods
Data collection
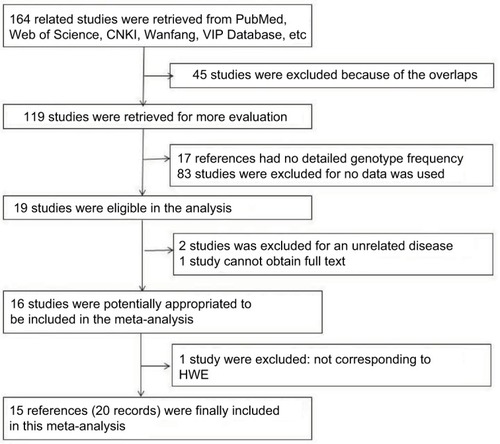
We searched related references from PubMed, Web of science, Chinese National Knowledge Infrastructure(CNKI), China Science and Technology Journal Database, and the Chinese Wanfang Data Knowledge Service Platform. The search keywords were “LncRNA AND cancer”, “CASC8”, “rs10505477”, and “rs7837328”. A total of 164 references were retrieved. After examination by title and abstract, 119 articles were retrieved for further evaluation. One hundred studies were excluded because of the absence of detailed genotype frequency, reviews, and cell line or animal studies. Two studies were excluded because of no specified cancer risk. One paper was removed because its full text could not be obtained. One study did not accord with the Hardy–Weinberg equilibrium (P=0.015) and, then, was excluded from the study. Finally, 15 articles containing 20 datasets (24,504 cases and 22,969 controls) were included and used in quantitative synthesis for systematic review. A flowchart of the study selection process is shown in .
Inclusion/exclusion criteria
Studies included in this meta-analysis had to meet the following criteria: 1) cases were diagnosed with carcinomas at any stage; 2) a clinical case–control study; 3) published in Chinese or English language; and that 4) the distribution of genotype in controls was consistent with the Hardy–Wein-berg equilibrium (HWE). Exclusion criteria were as follows: 1) comprehensive data on one type of cancer with other cancers were excluded in the stratified analyses by cancer types; 2) no definite genotype or allele frequency; 3) case–control studies that were family based; and 4) the document type was a summary or a review.
Statistical analysis
This meta-analysis describes the relationship between lncRNA CASC8 SNPs and various type of cancers with ORs and 95% CIs. For each SNP, we estimated five genetic models of ORs and 95% CIs, involving additive model, dominant model, and recessive model as well as homozygous and heterozygous comparisons. Subgroup analysis was conducted according to ethnicity, source of controls, genotyping methods, and cancer types.
The Hardy–Weinberg equilibrium test was conducted on the allele frequency of the control group. The study population were regarded as originating from the same Mendelian genetic group when P>0.05. The stability and effect of the results were assessed by the chi-square test as well as by calculating ORs and 95% CI. The combined ORs were calculated by the additive model (rs10505477 C vs T and rs7837328 A vs G), and the statistical significance was evaluated by the Z test. Cochran’s Q test and I2 were used to test the heterogeneity. If I2 <50% and P>0.1, the fixed effects model was used to calculate the ORs and the 95% CI; in contrast, the random effects model was applied. Publication bias was estimated by Begg funnel and Egger regression tests. All statistical analyses were conducted on STATA software (version 11.0), and P<0.05 of the two-tailed probability was considered to be statistically significant.
Results
Eligible studies
According to the inclusion/exclusion criteria, 15 articles containing 20 datasets (24,504 cases and 22,969 controls) were finally included in the meta-analysis. In these papers, nine articles including 14 records (16,238 cases and 16,594 controls) were related to rs10505477, and six studies including six datasets (8,266 cases and 6,375 controls) were con cerned with rs7837328. The characteristics of all studies are summarized in . The nine articles of rs10505477 were included in this meta-analysis.Citation11,Citation14,Citation16–Citation22 Among these, 11 records were Caucasian-based (13,652 cases and 14,221 controls), and three records were Asian-based (2,586 cases and 2,373 controls). In the same way, six articles about rs7837328 were selected in this study.Citation15,Citation23–Citation27 Caucasians had two records (1,341cases and 1,260 controls), whereas Asians had four records (6,925 cases and 5,115 controls).
Table 1 Characteristics of the studies included in this meta-analysis
Meta-analysis
Rs10505477 and cancer susceptibility
The relationship between the rs10505477 locus in the lncRNA CASC8 gene and the risk of all cancers is shown in . Compared to individuals carrying the TT genotype, those with the TC or CC genotype were at decreased risk of developing cancer (TC vs TT: OR 0.876, 95% CI 0.832–0.923, P<0.001; CC vs TT: OR 0.748, 95% CI 0.703–0.795, P<0.001). The same results were suggested in the dominant model (TC+CC vs TT: OR 0.834, 95% CI 0.794–0.875, P<0.001) and the recessive model (CC vs TC+TT: OR 0.817, 95% CI 0.776–0.860, P<0.001). In the additive model (C vs T), allele C might be a protective factor for decreasing the susceptibility to cancer (OR 0.866, 95% CI 0.840–0.893, P<0.001). The heterogeneity test is shown in (I2>50% and P>0.10), and the fixed effects model was used for the analyses.
Table 2 Relationship between SNP of rs10505477 and cancer susceptibility
The results of subgroup analysis are summarized in . In the ethnicity analysis, decreased cancer risk was found in either the Caucasian or Asian population. According to the source of controls, all the five models based on population-based studies showed statistical significance with regard to decreasing the risk of cancers (OR<1, P<0.001), whereas no significant difference was found in hospital-based studies (P>0.05). In studies using Taqman as the genotyping method, all models had a marked association with lower cancer susceptibility. For studies using the MassARRAY method, a significant relationship was found in homozygote comparison – recessive model and additive model. Significant correlations with decreased CRC risk were observed in all the models. Subject to the amount of included studies, other types of cancer have not been analyzed.
Table 3 The subgroup analysis of rs10505477 and cancer susceptibility
Rs7837328 and cancer susceptibility
Rs7837328 – another locus on lncRNA CASC8 – is associated with the risk of cancer susceptibility as shown in . Compared to the wild genotype (GG), the heterozygote genotype (GA) and the homozygote genotype (AA) were associated with increased risks of cancer (ORs were 1.209 and 1.336, 95% CIs were 1.127–1.298 and 1.202–1.484, respectively). Moreover, we drew the same conclusion in a dominant model (OR 1.236, 95% CI 1.156–1.322, P<0.001) and a recessive model (OR 1.204, 95% CI 1.092–1.328, P<0.001). In an additive model, allele A could significantly increase the risk of cancer as compared with allele G (OR 1.169, 95% CI 1.114–1.227, P<0.001). Heterogeneity test results are listed in ; owing to I2<50% and P>0.1, the fixed effects models were used.
Table 4 Relationship between SNP of rs7837328 and cancer susceptibility
Subgroup analysis was conducted by means of ethnicity, source of controls, genotyping methods, and cancer types. The results were revealed in . In different ethnicities, the additive, dominant, and genetic models of the Caucasian population had statistical significance in increasing cancer susceptibility (P<0.05). All models in the Asian population obtained a significant result in increasing the cancer risk (P<0.001). Significant results were found in all the genetic models for population-based studies (P<0.001). For the pooled results in hospital-based studies, in addition to a dominant model (P=0.151) and heterozygote comparison (P=0.489), other models showed a statistically significant effect on the cancer risk (P<0.05). In a subgroup analysis of cancer types, dominant models in CRC and pCa had a statistical association with increasing cancer susceptibility (P< 0.05). The additive model of pCa suggested a similar result (OR 1.147, 95% CI 1.045–1.258, P=0.004).
Table 5 The subgroup analysis of rs7837328 and cancer susceptibility
Influential analysis
In this study, we used the “influence analysis-metaninf ” method as a sensitivity analysis method to assess the reliability of this meta-analysis. After individually excluding studies, the combined effect of the OR value revealed no significant change; therefore, the results of this analysis are reliable.
Publication bias
Both Begg’s and Egger’s tests were used to estimate publication bias of rs10505477 and rs7837328 in this meta-analysis. Begg’s funnel plots of both polymorphisms were basically symmetrical; therefore, no publication bias was found in both loci. By Egger’ test, P-values were 0.061 and 0.746 for rs10505477 and rs7837328, respectively, but none of them had statistical significance.
Discussion
This is an updated meta-analysis on all available studies to assess the overall cancer risk with rs10505477 and rs7837328 polymorphisms in the CASC8 gene. The results showed that rs10505477 (T>C) and rs7837328 (G>A) polymorphisms were related to the risk of all kinds of cancers. In a subgroup analysis of cancer types, significant correlations of rs10505477 with decreased CRC risk were observed. Rs7837328 was suggested to be associated with the risks of CRC and pCa.
A malignant tumor that develops between the dentate line of the digestive tract and the sigmoid colon, CRC is the third most commonly diagnosed cancer in males and the second in females, with more than 1.2 million new patients and 608,700 deaths evaluated to have occurred in 2008.Citation28 pCa refers to epithelial malignancies that occur in the prostate. It was reported that pCa was the second most common cause of cancer in US men among non-cutaneous cancers.Citation29 Molecular epidemiology has increasingly shown that SNPs play a crucial role in the progression of cancer, including CRC and pCa.
In recent years, GWAS have authenticated five SNPs (rs6983267, rs10505477, rs7837328, rs10505477, and rs16892766) located in the 8q23–8q24 chromosome region, which had a strong correlation with the development of CRC.Citation13,Citation30–Citation32 SNPs, as third-generation genetic biomarkers, are widely distributed in the human genome and have good stability and high density. They can effectively reflect the differences of individuals to a certain extent, which has gradually become an important tool for medical research and molecular biology studies.
The mechanism by which the CASC8 gene modifies cancer susceptibility is still unknown. The possible role of CASC8 in cancer development is as follows: CASC8 is located near the MYC gene in the region of 8q24.1 – a known gene desert containing multiple enhancer elements in the proximity of the MYC gene, associated with several cancers, including pCa and CRC.Citation33 These enhancers regulate transcription of the MYC gene through an interaction with the CASC8 promoter.Citation5 The pathogenesis is speculated to be as follows: First, the rs10505477 allele could disrupt the correlation between CASC8 and the cognate gene POU5F1B (POU class 5 homeobox 1 pseudogene 1), whose carcinoma susceptibility is well known.Citation34 Thus, the mutant allele suppresses some transcription elements to act as the promoter of the POU5F1B gene.Citation14 Second, it was reported that a strong linkage disequilibrium (LD) was found between several loci (rs10505477,Citation35 rs7837328,Citation27 and rs7014346Citation36) and rs6983267, which is located at 8q24 and has confirmed to be related to CRC, pCa, and kidney cancer susceptibility, among others.Citation13,Citation37,Citation38 Resequencing and detailed determination of the regional LD indicated that rs6983267 could be a casual variation in disease. However, the variant is located in a gene desert.Citation39 The oncogene MYC, the proximal gene, is ~335 kb telomeres from the risk region, which is abnormally expressed in several cancers, including CRC.Citation40 Therefore, the rs10505477 and rs7837328 loci might indirectly affect the risk of CRC through their LD link with the cancer susceptibility-related rs6983267 locus.
Multiple polymorphisms in the 8q24 region have been proved to have a significant association with pCa risk that could be drawn in both case–control association and genetic linkage studies. Salinas et al confirmed that several 8q24 SNPs of western European descent and the centromeric- specific boundary of the 8q24 region are significantly associated with the risk of pCa, including rs6983267.Citation25
In an ethnicity subgroup analysis, we validated positive results in both Caucasian and Asian populations, wherein the aberrant expression of the mutant allele of rs10505477 indeed increased the risk of cancers. The ethnicity analysis of rs7837328 drew a significant conclusion in the Caucasian population, but negative results in the Asian population. The reason may be that although rs7837328 and the risk of CRC had been confirmed by GWAS, its correlation with pCa is still uncertain in Asians, particularly the Chinese. In southern Chinese, rs7837328 was reported to not be associated with PCa.Citation26 Interestingly, a statistically significant result was found in the northern Chinese Han population, for both of the alleles (P=0.004) and genotypes (P=0.008).Citation15 This discrepancy may be explained by daily lifestyle, dietary habits, geographic climate, ethnic diversity, and so on.
According to the relevant literature so far, in addition to another meta-analysis of 8q23-24-related loci and CRC in 2015,Citation41 this is the first study on the relationship between these loci and cancer susceptibility. Besides, our data are relatively new, with the latest data from 2016. In addition, we conducted a detailed subgroup analysis of ethnicity, source, genotyping, and cancer types, with high reliability.
The limitations of this study are as follows: first, the sample size was not large, and unpublished studies may exist that could introduce a potential publication bias; second, the research only studied the relationship between gene polymorphisms and cancer risks, ignoring environmental factors and the interaction of gene–environment; and third, due to data limitations, no tumor staging was discussed.
In conclusion, our meta-analysis showed that the rs10505477 (T>C) and rs7837328 (G>A) polymorphisms were related to the risk of cancer. Although limitations exist, future more rigorous studies are warranted to confirm this result.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant no. 81673261) and the Natural Science Foundation of Liaoning Province (grant no. 201602870).
Disclosure
The authors report no conflicts of interest in this work.
References
- BhanASoleimaniMMandalSSLong noncoding RNA and cancer: a new paradigmCancer Res201777153965398128701486
- BhanAMandalSSLong noncoding RNAs: emerging stars in gene regulation, epigenetics and human diseaseChemMedChem2014991932195624677606
- Martens-UzunovaESBöttcherRCroceCMJensterGVisakorpiTCalinGALong noncoding RNA in prostate, bladder, and kidney cancerEur Urol20146561140115124373479
- XiangJFYangLChenLLThe long noncoding RNA regulation at the MYC locusCurr Opin Genet Dev201533414826254776
- KimTCuiRJeonYJLong-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5Proc Natl Acad Sci U S A2014111114173417824594601
- MaGGuDLvCGenetic variant in 8q24 is associated with prognosis for gastric cancer in a Chinese populationJ Gastroenterol Hepatol201530468969525302443
- TanCHuWHuangYZhouJZhengSRisk of eighteen genome-wide association study-identified genetic variants for colorectal cancer and colorectal adenoma in Han ChineseOncotarget2016747776517766327769063
- BerndtSIPotterJDHazraAPooled analysis of genetic variation at chromosome 8q24 and colorectal neoplasia riskHum Mol Genet200817172665267218535017
- BeutenJGelfondJAMartinez-FierroMLAssociation of chromosome 8q variants with prostate cancer risk in Caucasian and Hispanic menCarcinogenesis20093081372137919528667
- LochheadPNgMTHoldGLPossible association between a genetic polymorphism at 8q24 and risk of upper gastrointestinal cancerEur J Cancer Prev2011201545721102338
- ZankeBWGreenwoodCMRangrejJGenome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24Nat Genet200739898999417618283
- HeJWilkensLRStramDOGeneralizability and epidemiologic characterization of eleven colorectal cancer GWAS hits in multiple populationsCancer Epidemiol Biomarkers Prev2011201708121071539
- TomlinsonIWebbECarvajal-CarmonaLA genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21Nat Genet200739898498817618284
- HuLChenSHLvQLClinical significance of long non-coding RNA CASC8 rs10505477 polymorphism in lung cancer susceptibility, platinum-based chemotherapy response, and toxicityInt J Environ Res Public Health2016136 pii: E545
- ZhangLLSunLZhuXQrs10505474 and rs7837328 at 8q24 cumulatively confer risk of prostate cancer in Northern Han ChineseAsian Pac J Cancer Prev20141573129313224815458
- HutterCMSlatteryMLDugganDJCharacterization of the association between 8q24 and colon cancer: gene-environment exploration and meta-analysisBMC Cancer20101067021129217
- SchafmayerCBuchSVölzkeHInvestigation of the colorectal cancer susceptibility region on chromosome 8q24.21 in a large German case-control sampleInt J Cancer20091241758018839428
- GruberSBMorenoVRozekLSGenetic variation in 8q24 associated with risk of colorectal cancerCancer Biol Ther2007671143114717630503
- ZhouCPPanHZLiFXHuNYLiMYangXXAssociation analysis of colorectal cancer susceptibility variants with gastric cancer in a Chinese Han populationGenet Mol Res20141323673368024854447
- HaerianMSHaerianBSRookiHAssociation of 8q24.21 rs10505477-rs6983267 haplotype and age at diagnosis of colorectal cancerAsian Pac J Cancer Prev201415136937424528058
- CurtinKLinWYGeorgeRMeta association of colorectal cancer confirms risk alleles at 8q24 and 18q21Cancer Epidemiol Biomarkers Prev200918261662119155440
- HashemiMBahariGNaderiMSadeghi BojdSTaheriMAssociation of lnc-LAMC2-1:1 rs2147578 and CASC8 rs10505477 polymorphisms with risk of childhood acute lymphoblastic leukemiaAsian Pac J Cancer Prev201617114985498928032728
- CuiROkadaYJangSGCommon variant in 6q26-q27 is associated with distal colon cancer in an Asian populationGut201160679980521242260
- San FranciscoIFRojasPATorres-EstayVAssociation of RNASEL and 8q24 variants with the presence and aggressiveness of hereditary and sporadic prostate cancer in a Hispanic populationJ Cell Mol Med201418112513324224612
- SalinasCAKwonECarlsonCSMultiple independent genetic variants in the 8q24 region are associated with prostate cancer riskCancer Epidemiol Biomarkers Prev20081751203121318483343
- ZhengSLHsingAWSunJAssociation of 17 prostate cancer susceptibility loci with prostate cancer risk in Chinese menProstate201070442543219866473
- YangBThyagarajanBGrossMDGoodmanMSunYVBostickRMGenetic variants at chromosome 8q24, colorectal epithelial cell proliferation, and risk for incident, sporadic colorectal adenomasMol Carcinog201453Suppl 1E187E19223776012
- JemalABrayFCenterMMFerlayJWardEFormanDGlobal cancer statisticsCA Cancer J Clin2011612699021296855
- SiegelRNaishadhamDJemalACancer statistics, 2012CA Cancer J Clin2012621102922237781
- ZhangBJiaWHMatsudaKLarge-scale genetic study in East Asians identifies six new loci associated with colorectal cancer riskNat Genet201446653354224836286
- WhiffinNHoskingFJFarringtonSMIdentification of susceptibility loci for colorectal cancer in a genome-wide meta-analysisHum Mol Genet201423174729473724737748
- TomlinsonIPWebbECarvajal-CarmonaLA genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3Nat Genet200840562363018372905
- SoteloJEspositoDDuhagonMALong-range enhancers on 8q24 regulate c-MycProc Natl Acad Sci U S A201010773001300520133699
- KastlerSHonoldLLuedekeMPOU5F1P1, a putative cancer susceptibility gene, is overexpressed in prostatic carcinomaProstate201070666667420017164
- HaerianMSBaumLHaerianBSAssociation of 8q24.21 loci with the risk of colorectal cancer: a systematic review and meta-analysisJ Gastroenterol Hepatol201126101475148421722176
- KupferSSAndersonJRHookerSGenetic heterogeneity in colorectal cancer associations between African and European AmericansGastroenterology20101395167716851685.e11685.e820659471
- DaraeiASalehiRSalehiMEffect of rs6983267 polymorphism in the 8q24 region and rs4444903 polymorphism in EGF gene on the risk of sporadic colorectal cancer in Iranian populationMed Oncol20122921044104921567271
- WokolorczykDGliniewiczBSikorskiAA range of cancers is associated with the rs6983267 marker on chromosome 8Cancer Res200868239982998619047180
- YeagerMXiaoNHayesRBComprehensive resequence analysis of a 136 kb region of human chromosome 8q24 associated with prostate and colon cancersHum Genet2008124216117018704501
- PomerantzMMAhmadiyehNJiaLThe 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancerNat Genet2009418882884
- LiLLvLLiangYAssociation of 8q23-24 region (8q23.3 loci and 8q24.21 loci) with susceptibility to colorectal cancer: a systematic and updated meta-analysisInt J Clin Exp Med2015811210012101326885031