Abstract
Purpose
Smurf2 is a member of the homologous to E6-AP carboxyl terminus family of E3 ubiquitin ligases. Changes in their expression pattern are known to contribute to tumorigenesis. Smurf2 plays a decisive role in cell differentiation, proliferation, and migration and exhibits a dual role in cancer – functioning as both oncogene and tumor suppressor. Dysregulation of Smurf2 in different cancer types has been described, besides colorectal cancer (CRC). We therefore examined the expression and oncogenic potential of Smurf2 in human CRC patients.
Materials and methods
Expression levels of Smurf2 were analyzed via qRT-PCR in CRC specimens and healthy mucosa from 98 patients who had undergone surgery due to CRC. Spatial expression of Smurf2 was additionally studied by immunohistochemistry. siRNA-mediated knockdown of Smurf2 was applied for migration and invasion assays in DLD-1 and SW-480 cells.
Results
Smurf2 was significantly overexpressed in CRC tissue compared to corresponding healthy colon mucosa. Smurf2 expression levels differed significantly between microsatellite instable (MSI) and microsatellite stable (MSS) CRC. In patients suffering from MSS CRC, high tumoral expression of Smurf2 was significantly associated with impaired overall survival. Consistently, in vitro analysis revealed that knockdown of Smurf2 reduced the invasive and migratory potential of MSS CRC cells.
Conclusion
Smurf2 expression is upregulated in CRC specimens and affects survival dependent on patients’ MSI status. Moreover, Smurf2 supports cancer cell migration and invasion, collectively suggesting an oncogenic function in CRC.
Introduction
The ubiquitin–proteasome system (UPS) is required for protein degradation through protein ubiquitination.Citation1 Protein ubiquitination by the UPS plays a substantial role in a variety of cellular processes such as differentiation, proliferation, DNA repair, signal transduction and transcription, angiogenesis, and apoptosis.Citation2–Citation4 The UPS consists of ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2, and a substrate-specific E3 ubiquitin ligase.Citation1 Dysregulation in the expression pattern of the substrate-specific E3 ubiquitin ligases is known to contribute to tumorigenesis.Citation1 Smad ubiquitination regulatory factor 2 (Smurf2) belongs to the homologous to E6-AP carboxyl terminus (HECT) family of E3 ubiquitin ligases.Citation5 It displays a contradictory role in cancer exhibiting both tumorsuppressive and oncogenic functions.Citation2,Citation6,Citation7 Tumor suppressive functions of Smurf2 are linked to strengthening of p53 and p21, resulting in cell cycle senescence.Citation3 By contrast, Smurf2 leads to tumor progression and promotes metastases via upregulation of epithelial-to-mesenchymal transition markers such as N-cadherin.Citation3 Moreover, Smurf2 inhibits apoptosis due to mouse double minute 2 homolog-(MDM2) mediated inhibition of p53.Citation8 Importantly, as a member of the UPS, E3 ligase Smurf2 leads – along with its binding partner Smad7 – to TGF-ß receptor degradation. It modulates TGF-ß pathway signaling by regulating the degradation of Smad1, Smad2, and Smad3, thus promoting cancer progression via altered TGF-ß signaling.Citation5,Citation9,Citation10
Altered expression of Smurf2 has been observed in a variety of solid cancer types, such as breast or esophageal cancer.Citation7,Citation11,Citation12 Given its function in the above mentioned signaling pathways, it is not astonishing that Smurf2 silencing diminishes the invasion and migration of breast cancer cells.Citation12 Besides, high-level expressional changes of Smurf2 are associated with poor prognosis in esophageal cancer.Citation11 Nonetheless, until now, no data exist regarding Smurf2 expression and its oncogenic relevance in CRC patients. Therefore, we examined Smurf2 expression levels in a collective of 98 CRC patients, comparing colorectal carcinoma tissue with corresponding healthy mucosa. Subsequently, Smurf2 expression levels were compared with clinical data including overall survival. The impact of Smurf2 expression on cancer cell migration and invasion was additionally assessed applying siRNA-mediated knockdown in DLD-1 and SW-480 CRC cells.
Materials and methods
Patients and tissue samples
Ninety-eight patients suffering from colorectal cancer (CRC) who underwent surgery at the Department of General, Visceral and Transplantation Surgery, University of Heidelberg, Germany, between 2007 and 2013 were included. Fresh frozen tissue specimens of healthy mucosa and colorectal carcinomas were used for qRT-PCR. Healthy mucosa tissue was sampled at least 10 cm distant from the tumor. Formalin-fixed paraffin-embedded tissue samples of colorectal carcinoma and corresponding healthy colon mucosa were provided from the tissue bank of the National Center for Tumor Disease, University of Heidelberg, Germany for immunohistochemical staining. Every patient gave written informed consent and the local ethics committee of the University Heidelberg approved the study. Clinical and histopathological characteristics including date of birth, gender, tumor location, TNM classification, UICC (Union internationale contre le cancer) stage, R classification, grading, neoadjuvant radiochemotherapy, adjuvant chemotherapy, microsatellite stability (MSS) status, postoperative complications, and overall survival (time from operation up to death or last follow-up) were obtained from each patient.
RNA isolation and qRT-PCR
Total RNA from tumor tissue and corresponding healthy mucosa was isolated using RNeasy® Mini Kit (Qiagen, Hilden, Germany) and reverse-transcribed using ImProm-II™ Reverse Transcription System (Promega, Mannheim, Germany) according to the manufacturer’s instructions. Resulting cDNA was amplified and detected with LightCycler® 480 SYBR Green I Master (Roche Diagnostics GmbH, Mannheim, Germany) in a Roche Light Cycler® 480 System (Roche Diagnostics GmbH). Predesigned primers of Smurf2 (HS_SMURF2_1_SG QuantiTect Primer Assay) and 18S (HS_RRN18S_1_SG QuantiTect Primer Assay) were purchased from Qiagen (sequences available on request).
Immunohistochemistry
Slides were pretreated at pH 9.0, and immunohistochemical staining of 5 µm thick formalin-fixed paraffin-embedded slides of CRC tissue and corresponding healthy mucosa for Smurf2 (Anti-Smurf2 ab38543, Abcam, Cambridge, UK) was performed using the Dako autostainer with 1:200 dilution. A board certified pathologist from the Institute of Pathology, University of Heidelberg performed histopathological assessment.
Cell culture and transfection
DLD-1 (ATCC® CCL-221™, LGC Standards GmbH, Wesel, Germany) and SW480 (ATCC® CCL-228™, LGC Standards GmbH) CRC cells were used for in vitro studies. Both cell lines were cultured in RPMI-1640 (Sigma-Aldrich GmbH, Munich, Germany), supplemented with 10% FBS (Gibco® by Life Technologies, Darmstadt, Germany), 100 U/mL penicillin, and 100 µg/mL streptomycin (Sigma-Aldrich GmbH) at 37°C and 5% CO2.
Smurf2 siRNA (Stealth™ RNAi Smurf2 HSS127687) and scrambled siRNA as a control nonsilencing sequence (Stealth™ RNAi Negative Control, both from Life Technologies GmbH, Darmstadt, Germany) were transfected using Lipofectamine® RNAiMAX (Life Technologies GmbH) according to the manufacturer’s protocol. Transfection efficiency was monitored with semiquantitative RT-PCR and calculated as the difference between siControl and the residual Smurf2 expression after transfection for each invasion and migration assay separately.
Cell migration and invasion assays
Twenty-four hours after transfection, DLD-1 and SW-480 cells were harvested and starved in serum-free medium prior to experiments. Migration and invasion assays were performed using ThinCerts™ (24 wells, pore size 8.0 µm, Greiner Bio-One, Frickenhausen, Germany) and BD BioCoat™ Matrigel™ Invasion Chambers (Greiner Bio-One), respectively, according to manufacturer’s protocol. Incubation time was 24 hours for cell migration and 72 hours for cell invasion assay. Readouts were determined by spectrophotometric readings on a microtiter plate at 540 nm with a microplate reader. Both assays were performed in triplicate.
Statistics
Statistical analyses were conducted with Excel 2013 (Microsoft Corporation, Redmond, WA, USA) and SPSS, version 22 (SPSS, IBM Corporation, Armonk, NY, USA). Paired Student’s t-test was used to determine expressional differences in different tissues. Expressional data are presented as mean + standard error of the mean. Expressional differences of Smurf2 in MSS vs microsatellite instable (MSI) tumors were assessed using Mann–Whitney U-test. Statistical differences of clinical parameters such as T stage, N stage, grading, resection margin, tumor site, surgical procedure, and localization of recurrence were assessed with Kruskal–Wallis test. Statistical differences of clinical parameters such as gender, age, M stage, liver metastases, neoadjuvant chemotherapy, anastomotic leakage, microsatellite stage, recurrence, and primary stoma were assessed using Mann–Whitney U-test. Median follow-up time was calculated as median difference between time of operation and last follow-up or death of patients. Kaplan–Meier method was employed to estimate cancer-related overall survival. Differences between survival curves were evaluated by log-rank test. Results were considered significant at a P-value <0.05.
Results
Patients and clinical data
In total, 98 patients who underwent surgery due to colorectal adenocarcinoma were included in the study. Median age at the time of operation was 64 years. Mean overall survival was 55.5 months. Median follow-up time was 619 days. Twenty patients died during the follow-up. A detailed overview on patients’ clinical characteristics is provided in .
Table 1 Overview of clinical parameters and expression of Smurf2
Quantitative expression of Smurf2 in colorectal cancer tissue and healthy mucosa
Real-time PCR of CRC tissue and corresponding healthy mucosa revealed a significantly higher expression of Smurf2 in tumoral tissue than in healthy mucosa (P =0.003) (). We then correlated Smurf2 expression with patients’ clinical data (). Interestingly, MSS (n=58) tumors displayed significantly lower expression of Smurf2 compared to MSI (n =11) tumors (P =0.006) (). No statistically significant correlations with tumoral Smurf2 expression were observed regarding other histopathological or clinical variables ().
Figure 1 Transcript expression levels of Smurf2 in colorectal cancer tissue and corresponding healthy mucosa (n=98).
Notes: Smurf2 was significantly overexpressed in colorectal cancer specimens compared to corresponding healthy mucosa. Bars represent mean + SEM. **P=0.003.
Abbreviation: SEM, standard error of the mean.
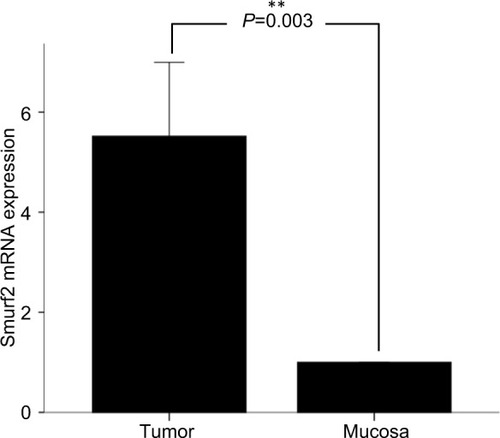
Figure 2 (A) Smurf2 expression dependent on patients’ microsatellite status. Patients with microsatellite stable (MSS) tumors (n=58) displayed a significantly lower expression of Smurf2 than patients with microsatellite instable (MSI) tumors (n=11; P=0.006). (B) Within the 58 MSS tumors, Smurf2 expression was significantly higher in M1 staged patients vs M0 (P=0.024).
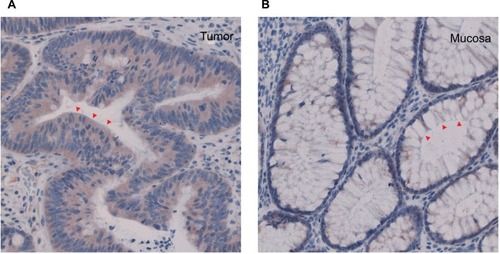
Spatial expression of Smurf2 in colorectal cancer tissue and healthy mucosa
Immunohistochemical staining was performed in five patients in order to assess cellular localization of Smurf2 in CRC tissue and normal colon mucosa. Consistent with the quantitative expression analysis outlined above, Smurf2 was strongly expressed in the tumor cells, particularly in the cytoplasm, whereas healthy mucosa cells only showed a very weak expression of Smurf 2 ().
Prognostic impact
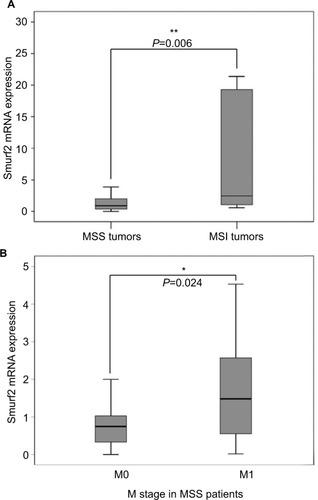
Subsequently, we correlated tumoral Smurf2 expression data with patients’ overall survival. For this purpose, patients were divided into two groups: patients with high tumoral Smurf2 expression (an expression of Smurf2 five times higher in the tumoral tissue than in corresponding healthy mucosa) and patients with low tumoral Smurf2 expression (an expression of Smurf2 less than five times in the tumoral tissue than in corresponding healthy mucosa). In the entire collective of patients (n=98), mean overall survival was 55.5 months. When looking at the entire cohort, mean overall survival was 58 months for patients displaying high Smurf2 expression, compared to 52.9 months in patients with low tumoral Smurf2 expression (n=98) (P=0.86) (). Interestingly, in the subgroup of patients with MSS tumors (n=58), high tumoral expression of Smurf2 was significantly associated with impaired overall survival (mean overall survival high vs low Smurf2 expression 38.3 vs 67.4 months; P=0.044) (). By contrast, correlation of tumoral Smurf2 expression in patients with MSI tumors showed no influence on overall survival (n=11, P =0.41). Subgroup analysis of MSS tumors revealed a significant two times higher median tumoral Smurf2 expression in M1 vs M0 staged patients (n=58, P=0.024; , ).
Figure 4 (A) Correlation of Smurf2 expression and overall survival of the entire patient cohort revealed no significant difference between Smurf2 high and Smurf2 low tumors (n=98) (P=0.86). (B) Correlation of Smurf2 with microsatellite status and overall survival. Overall survival was significantly impaired in microsatellite stable (MSS) patients expressing high levels of Smurf2 (n=58) (*P=0.044).
Influence of Smurf2 on migration and invasion of SW-480 and DLD-1 cells in vitro
In vitro assays were conducted in order to assess the impact of Smurf2 expression on the invasive and migrative potential of CRC cells. For this purpose, we chose a MSS CRC cell line (SW-480) as well as a MSI cell line (DLD-1). Upon knockdown of Smurf2, we observed significantly diminished invasive (P=0.012) and migrative (P=0.011) properties of MSS SW-480 cells (). MSI DLD-1 cells likewise displayed significantly reduced invasive potential (P=0.013), but only insignificantly reduced migrative potential upon Smurf2 knockdown (P=0.11) (). The mean transfection efficiency was 76% (range 73%–79%) in SW-480 cells and 64.3% (range 52%–84%) in DLD-1 cells for the migration assays. For the invasion assays, mean transfection efficiency was 81.3% (range 73%–92%) in SW-480 cells and 85.3% (range 84%–88%) in DLD-1 cells.
Figure 5 Invasion (A) and migration (B) assays.
Notes: Smurf2 siRNA-transfected SW-480 cells were both significantly less invasive (*P=0.012) and migrative (*P=0.011); DLD-1 cells revealed significantly reduced invasive potential (*P=0.013). Bars represent mean + SEM.
Abbreviation: SEM, standard error of the mean.
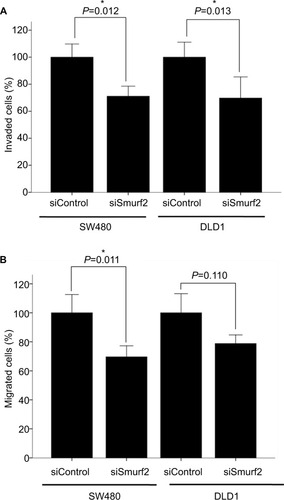
Collectively, these findings suggest that Smurf2 expression enhances the invasive and migrative properties of CRC cells.
Discussion
To our knowledge, this is the first study evaluating the prognostic impact of the E3 ubiquitin ligase Smurf2 in CRC patients. We detected significantly enhanced expression of Smurf2 in CRC tissue compared to corresponding healthy colon mucosa, as well as significantly attenuated expression of Smurf2 in MSS compared to MSI tumors. Intriguingly, high-level Smurf2 expression represented an adverse prognostic factor in patients with MSS tumors. In vitro, interference with Smurf2 expression was associated with impaired migrative and invasive proficiency of CRC cells. These findings indicate that Smurf2 could be a prognostic marker in MSS CRC patients.
Targeted degradation of proteins by the UPS is crucial for diverse biological processes such as cell-cycle progression, apoptosis, angiogenesis, stem cell quiescence, signal transduction, and transcriptional regulation.Citation13,Citation14 Expressional changes, dysfunction, and genetic alterations of substrate-specific E3 ubiquitin ligases are known to provoke cancer.Citation2 The E3 ubiquitin ligase Smurf2 is located on chromosome 17 and exhibits, contrary to Smurf1, only one isoform. It possesses a C2 domain, a C-terminal HECT domain, and three central WW protein-interacting domains, of which second and third are responsible for target recognition.Citation3 Dependently on its particular targets, Smurf2 can induce different signaling pathways – either oncogenic or tumorsuppressive.
On the cellular level, Smurf2 is able to transfer between the nucleus and the cytoplasm: after interacting with Smad6/Smad7 inside the nucleus, the Smurf2/Smad complex shuttles to the cytoplasm to target specific substrates.Citation3 Immunohistochemical staining applied in our present study revealed expression of Smurf2 protein predominantly in the cytoplasm of CRC cells, indicating activated complex formation enabling further substrate binding and active signaling. Importantly, quantitative expression analysis revealed overexpression of Smurf2 in CRC tissue compared to healthy colon mucosa. This is consistent with some previous reports on Smurf2 expression in solid cancers: For instance, Fukuchi et al described overexpression of Smurf2 in esophageal squamous cell carcinoma.Citation11 By contrast, downregulation of Smurf2 is a hallmark of triple-negative breast cancers – but not of hormone receptor-positive breast cancers.Citation15 Hellwinkel et al reported higher Smurf2 expression in T2 than in T3 prostate cancer, indicating that loss of Smurf2 facilitates tumor progression.Citation16 In concert with these previous reports, our present findings emphasize on putatively dual role of Smurf2 in cancer progression, which is most likely due to activation of different signaling pathways in various tumor entities and individual patient settings.
Our current in vitro analysis links expressional downregulation of Smurf2 to diminished migrative and invasive potential of CRC cells. This phenomenon was independently observed in two established CRC cell lines and pronounced in the MSS SW-480 cell line. It is thus conceivable that Smurf2 facilitates metastatic properties of CRC cells, at least in MSS CRC. In accordance with these findings, David et al observed attenuated proliferative, migrative, and invasive properties after Smurf2 knockdown in MCF-7 (estrogen receptor [ER] positive) and MDA-MB-231 (ER negative) breast cancer cells, which was mediated by the Smurf2 target protein connector enhancer of kinase suppressor of ras 2 (CNKSR2) via PI3K–PTEN–AKT signaling.Citation7 Moreover, Smurf2 reportedly fosters migration and invasion of breast cancer cells through upregulation of the epithelial–mesenchymal transition marker N-cadherin.Citation12 Consistent with these pro-oncogenic functions of Smurf2, Fukuchi et al associated high-level expression of Smurf2 with increased invasion and lymph node metastases in esophageal squamous cell carcinoma.Citation11
In our present study, we did not detect an association between Smurf2 expression in human CRC tissues and clinical TNM stages. However, Smurf2 expression was significantly elevated and associated with a wider range in MSI as compared to MSS tumors, hinting the fact that MSI tumors of CRC with DNA replication errors and defective mismatch repair have distinct phenotypic features in contrast to MSS tumors.Citation17 A relevant proportion of about 15% of CRC display microsatellite instability due to deficient DNA mismatch repair. About 3% of those are associated with a hereditary context such as Lynch syndrome, whereas 12% occur as sporadic CRCs.Citation18–Citation20 MSI/MSS status by itself is a predictor of prognosis in CRC patients,Citation21 albeit its prognostic impact varies.Citation22–Citation24 In general, better prognosis is reported in patients with MSI compared to those with MSS CRC,Citation22 but conflicting evidence likewise exists: Shin et al identified MSI as an independent prognostic factor, associated with impaired survival in stage II CRC,Citation23 and Mohan et al reported reduced disease-specific survival in stage III MSI CRC.Citation25 Given these discordant observations, it appears conceivable that the prognostic impact of patients’ MSI status varies, dependently on the expression of alternative tumoral signaling pathways. Remarkably, in this context, we found that high tumoral expression of Smurf2 was associated with impaired survival in patients with MSS CRC, but not in those with MSI CRC. Second, within MSS CRC, a significant elevation of Smurf2 expression was found in M1 staged patients. This can be explained as the decrease in invasion of the MSI cell line DLD-1 might correlate with patients T stage as a surrogate for invasiveness, but in vivo, the T stage did not show differences in Smurf2 expression (P =0.682, ). Furthermore, migration of the MSI cell line DLD-1 showed no significant difference after Smurf2 knockdown whereas the MSS cell line SW-480 showed both significant differences in invasion and migration. In general, locally advanced CRC correlate with impaired overall survival, but in our data, no influence on Smurf2 expression within different T stages in MSI CRC could be proven. This could be due to the small number of deaths in subgroup analysis, which is a potential weakness of our analysis. Another possibility is that invasiveness in vivo is regulated in a much more complex manner and Smurf2 expression alone cannot represent this adequately for MSI tumors. However, the influence of Smurf2 in MSS CRC on M stage and overall survival is consistent with our in vitro data outlined above, which revealed significantly reduced invasion and migration upon expressional silencing of Smurf2 particularly in an MSS CRC cell line.
Though the oncogenic potential of the E3 ubiquitin ligase Smurf2 has been proven,Citation3 studies linking the impact of Smurf2 expression to overall survival in individual tumor entities have remained scarce. Collectively, the results outlined in this study indicate a prognostic role of Smurf2 in MSS CRC patients. However, further studies are necessary to evaluate the oncogenic and pro-metastatic properties of Smurf2 dependently on patients’ microsatellite status as well as underlying molecular mechanisms.
Conclusion
We suggest an oncogenic role of E3 ubiquitin ligase Smurf2 in MSS CRC. Moreover, upregulated Smurf2 affects survival – dependent on patients’ MSI status – indicating its prognostic impact.
Author contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Acknowledgments
This work was supported by a grant from the Clinical Research Unit KFO 227: “From primary tumor progression towards metastases” funded by the German Research foundation (DFG).
Disclosure
The authors report no conflicts of interest in this work. The abstract of this paper titled “Smurf2 – an invasive and oncogenic E3-ubiquitin ligase in colorectal cancer” was presented at the 20th Surgical Research Days, Section of Surgical Research of the German Society of Surgery, September 2016, Magdeburg, Germany as a conference talk with interim findings. The abstract only was published in European Surgical Research, 2016¸Vol. 57; p.288; DOI 10.1159/000448816.
References
- NakayamaKINakayamaKUbiquitin ligases: cell-cycle control and cancerNat Rev Cancer20066536938116633365
- BernassolaFKarinMCiechanoverAMelinoGThe HECT family of E3 ubiquitin ligases: multiple players in cancer developmentCancer Cell2008141102118598940
- DavidDNairSAPillaiMRSmurf E3 ubiquitin ligases at the cross roads of oncogenesis and tumor suppressionBiochim Biophys Acta183520131119128
- ZhangYChangCGehlingDJHemmati-BrivanlouADerynckRRegulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligaseProc Natl Acad Sci USA200198397497911158580
- KavsakPRasmussenRKCausingCGSmad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradationMol Cell2000661365137511163210
- BlankMTangYYamashitaMBurkettSSChengSYZhangYEA tumor suppressor function of Smurf2 associated with controlling chromatin landscape and genome stability through RNF20Nat Med201218222723422231558
- DavidDJagadeeshanSHariharanRNairASPillaiRMSmurf2 E3 ubiquitin ligase modulates proliferation and invasiveness of breast cancer cells in a CNKSR2 dependent mannerCell Div20149225191523
- NieJXiePLiuLSmad ubiquitylation regulatory factor 1/2(Smurf1/2) promotes p53 degradation by stabilizing the E3 ligase MDM2J Biol Chem201028530228182283020484049
- BonniSWangHRCausingCGTGF-beta induces assembly of a Smad2 –Smurf2 ubiquitin ligase complex that targets SnoN for degradationNat Cell Biol20013658759511389444
- XuYPascheBTGF-beta signaling alterations and susceptibility to colorectal cancerHum Mol Genet200716Spec 1R14R2017613544
- FukuchiMFukaiYMasudaNHigh-level expression of the Smad ubiquitin ligase Smurf2 correlates with poor prognosis in patients with esophageal squamous cell carcinomaCancer Res200262247162716512499250
- JinCYangYAAnverMRMorrisNWangXZhangYESmad ubiquitination regulatory factor 2 promotes metastasis of breast cancer cells by enhancing migration and invasivenessCancer Res200969373574019155312
- InghamRJGishGPawsonTThe Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architectureOncogene200423111972198415021885
- InoueYImamuraTRegulation of TGF- beta family signaling by E3 ubiquitin ligasesCancer Sci200899112107211218808420
- SubramaniamVLiHWongMThe RING-H2 protein RNF11 is overexpressed in breast cancer and is a target of Smurf2 E3 ligaseBr J Cancer20038981538154414562029
- HellwinkelOJAsongLERogmannJPTranscription alterations of members of the ubiquitin-proteasome network in prostate carcinomaProstate Cancer Prostatic Dis2011141384521102547
- KimHJenJVogelsteinBHamiltonSRClinical and pathological characteristics of sporadic colorectal carcinomas with DNA replication errors in microsatellite sequencesAm J Pathol199414511481568030745
- BolandCRGoelAMicrosatellite instability in colorectal cancerGastroenterology2010138620732087.e320420947
- JaspersonKWTuohyTMNeklasonDWBurtRWHereditary and familial colon cancerGastroenterology201013862044205820420945
- AhadovaAGallonRGebertJThree molecular pathways model colorectal carcinogenesis in Lynch syndromeInt J Cancer2018143113915029424427
- SinicropeFASargentDJMolecular pathways: microsatellite instability in colorectal cancer: prognostic, predictive, and therapeutic implicationsClin Cancer Res20121861506151222302899
- PopatSHubnerRHoulstonRSSystematic review of microsatellite instability and colorectal cancer prognosisJ Clin Oncol200523360961815659508
- ShinUSChoSSMoonSMIs microsatellite instability really a good prognostic factor of colorectal cancer?Ann Coloproctol2014301283424639968
- WebberEMKauffmanTLO’ConnorEGoddardKASystematic review of the predictive effect of MSI status in colorectal cancer patients undergoing 5FU-based chemotherapyBMC Cancer20151515625884995
- MohanHMRyanEBalasubramanianIMicrosatellite instability is associated with reduced disease specific survival in stage III colon cancerEur J Surg Oncol201642111680168627370895