Abstract
Background
Hepatocellular carcinoma (HCC) is the third leading cause of death by malignancy worldwide. HCC has a poor prognosis due to tumor invasiveness and metastasis. There is substantial evidence that the epithelial-to-mesenchymal transition (EMT) plays a central role in cancer metastasis. In a previous study, a possible association between integrator complex 8 (INTS8) and the progression and development of HCC was discovered. However, its role and the molecular mechanisms in HCC are poorly understood.
Methods
The PROGgeneV2 platform database and Kaplan–Meier plotter analysis were used to analyze the potential effects of INTS8 in HCC. Moreover, we performed migration, transwell, and metastasis assays to investigate the effects of INTS8 on HCC cells. In addition, relevant signaling pathways were examined by western blot and RT-qPCR assays.
Results
We used the PROGgeneV2 platform database and Kaplan–Meier plotter analysis, which indicated that increased expression of INTS8 is associated with poor overall survival of HCC. Moreover, INTS8 expression was higher in HCC tissues than in adjacent noncancerous tissues. INTS8 depletion reduced the invasion and migration of HCC cell lines. Downregulation of INTS8 in vivo resulted in fewer observed metastatic nodules in lungs. Moreover, INTS8 knockdown also increased the expression of epithelial markers (E-cadherin) and decreased the expression of mesenchymal markers (N-cadherin and vimentin) following the downregulation of SMAD4. In addition, pretreatment with TGF-β1 could partly prevent the decrease in the expression of SMAD4 and EMT markers induced by INTS8 knockdown.
Conclusion
Overall, these findings suggest that INTS8 accelerates the EMT in HCC by upregulating the TGF-β signaling pathway.
Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of death from cancer.Citation1 The average and 5-year survival rates of HCC patients still remain poor. Its high recurrence and metastasis rates often lead to poor clinical outcomes.Citation2 Consequently, it is important to gain a better understanding of the mechanisms responsible for HCC initiation and progression, so that more effective therapies can be developed.
The integrator complex (INT) includes at least 14 subunits and is phylogenetically conserved.Citation3 INTS8 encodes a subunit of the INT that is involved in the cleavage of small nuclear RNAs. In a previous study, INTS8 levels were found to be significantly higher in cancerous gastric tissues than in the corresponding non-malignant gastric tissues.Citation4 Moreover, Simpson et al identified recurrent mutations in the INTS8 gene of peripheral T cell lymphoma.Citation5 The expression of INTS8 is robustly increased in cholangiocarcinoma, and colon and lung adenocarcinomas, as well as HCC. In addition, Yin et al also found that a significant increase at the protein level of INTS8 was observed in HCC tissues compared with corresponding paracancerous tissues, and INTS8 was specifically correlated with most clinical characteristics including disease-free survival, overall survival, stage, metastasis, invasiveness, diagnosis, and age by microarray analysis.Citation6
The epithelial-to-mesenchymal transition (EMT) is a multistep biological process whereby transient de-differentiation into a mesenchymal phenotype causes changes in the plasticity of epithelial cells.Citation7 The EMT, which is a reversible and dynamic process, is one of the major mechanisms by which cancer cells invade and metastasize.Citation8,Citation9 Accumulating evidence has demonstrated that the EMT plays a pivotal role in the proliferation and metastasis of malignant hepatocytes during hepatoma progression.Citation2,Citation10 Xiao et al revealed that HCC cell proliferation was inhibited through the downregulation in expression of stem cell markers.Citation11 Liang et al also reported that the inhibition of EMT decreased the cell proliferation in HCC.Citation12 TGF-β plays an important role in orchestrating favorable microenvironments for tumor cell growth and promoting the EMT in HCC.Citation13 Thus, we could assume that INTS8 might involve the EMT in HCC due to a possible association between INTS8 and the progression and development of HCC.
Strikingly, to date, the role of INTS8 in HCC and its underlying molecular mechanism remain unclear; thus, the aim of this study was to explore both.
Materials and methods
Bioinformatics analysis
The ProgeneV2 prognostic database (http://watson.compbio.iupui.edu/chirayu/proggene/database/?url=proggene) and Kaplan–Meier Plotter (http://kmplot.com/analysis/) were used to collect the data required to analyze the effect of INTS8 on HCC survival.Citation14,Citation15
Patients and clinical samples
This study was approved by the Medical Ethics Committee of the Ruijin Hospital of Shanghai Jiao Tong University School of Medicine and was conducted in accordance with the Declaration of Helsinki. We obtained written informed consent from all 52 participants, who were primary HCC patients enrolled between February 2015 and November 2016. We collected primary HCC tissues and corresponding adjacent noncancerous tissues during hepatectomies. Detailed clinical characteristics of the HCC patients are summarized in . The samples were confirmed as HCC and adjacent noncancerous tissues by H&E staining.
Table 1 Clinical characteristics of the HCC patients
Immunohistochemistry (IHC)
IHC staining was performed blindly by a pathologist. We assessed INTS8 expression by IHC of paraffin-embedded specimens. The slides were incubated overnight at 4°C with rabbit anti-INTS8 primary antibody (1:200; Proteintech, Wuhan, China). The following day, secondary antibody was applied for 30 minutes at 37°C and expression was quantified using ImageJ software.
Cell culture and stimulation
We purchased commercially available immortalized human hepatocyte (MIHA and THLE3) and liver cancer cell lines (HCCLM3, HepG2, MHCC-97H, and SK-HEP-1) from the American Type Culture Collection (Manassas, VA, USA). The cell lines were cultured in DMEM (Sigma-Aldrich Co., Shanghai, China) supplemented with 10% FBS (Invitrogen Gibco, Carlsbad, CA, USA) and incubated in a 5% CO2 incubator at 37°C. We prepared a 10 mg/mL stock solution by dissolving TGF-β1 (Abcam, Burlingame, CA, USA) in PBS and added sufficient medium to yield a 10 ng/mL solution.
Cell viability assay
The cells were seeded into 96-well plates and cultured to 60%–80% confluence. An MTT assay (Thermo Fisher Scientific, Waltham, MA, USA) was performed to determine cell viability and the absorbance was measured at 570 nm.
Reverse-transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol reagent (Invitrogen). We synthesized first strand cDNA using a reverse transcription system kit (Toyobo, Osaka, Japan) according to the manufacturer’s instructions. Real-time RT-PCR was performed using an ABI PRISM 7300 sequence detection system (Applied Biosystems, Foster City, CA, USA). All gene transcripts were quantified by RT-qPCR with the Power SYBR Green PCR Master Mix and the ABI StepOnePlus System. The forward primer sequences for INTS8 and β-actin, the internal control, were 5′-AACTGAGAGTTCTACTGCTGGA-3′ and 5′-GCTGCGCCCAAATCATAGC-3′, respectively, and the corresponding reverse sequences were 5′-GACTGCTGTCACCTTCACCGT-3′ and 5′-GTTGCGTTACACCCTTTCTTG-3′, respectively. We applied the 2−DDCt method to calculate relative gene expression.
Western blot (WB) analysis
We extracted total protein from the cell lines and tissues using radioimmunoprecipitation buffer (Sigma-Aldrich Co.) and determined the protein concentration using the BCA Protein Assay Kit (Beyotime, Shanghai, China). The extraction of nuclear protein extractions were prepared using an extraction kit (Beyotime) according to the manufacturer’s protocols as reported previously.Citation16 The proteins were separated by 10% SDS-PAGE and transferred to a polyvinylidene fluoride membrane. After blocking in 5% BSA for 1 hour, the membranes were incubated overnight at 4°C with β-actin (1:2,000; Cell Signaling Technology, Danvers, MA, USA), INTS8 (1:200; Abcam), N-cadherin (1:1,000; Abcam), E-cadherin (1:1,000; Abcam), Vimentin (1:1,000; Abcam), Histone H3 (1:2,000; Cell Signaling Technology), and SMAD4 (1:1,000; Cell Signaling Technology) antibodies. The secondary antibody horseradish peroxidase-conjugated goat anti-rabbit IgG (1:1,000) was then applied for 1 hour at 37°C. β-Actin served as the internal control for all WBs. Histone H3 served as the internal control for nuclear protein. We quantified protein expression using Bio-Rad Quantity One software (Bio-Rad Laboratories Inc., Hercules, CA, USA).
Lentiviral infection
We obtained the lentiviral shRNA construct targeting INTS8 (SHCLNV-NM_017864) commercially (Sigma-Aldrich Co.) and designed two shRNA sequences that interfere with human INTS8 (). The oligonucleotides were phosphorylated, annealed, and cloned into the pLKO.1 vector (Sigma-Aldrich Co.). Lentiviral infection was performed according to the manufacturer’s instructions. First, the cells were incubated with lentiviral particles and polybrene (1 µg/mL) in a growth medium. After 18 hours, the infection medium was discarded. After 48 hours, transfected cells were selected for 7 days with 1 µg/mL puromycin to produce stable INTS8-knockdown cell lines. INTS8 expression was quantified by RT-qPCR and WB analyses. The stable transfected cells were then used in subsequent experiments.
Table 2 shRNA sequences interfering with human INTS8
Colony formation assay
Colony formation assay was performed according to previous study.Citation17 Briefly, after lentivirus infection for 3 days, cells were seeded in a six-well plate at a density of 400 cells per well. Cells were cultured for 10 days at 37°C in a 5% CO2. The colonies were fixed with 4% paraformaldehyde for 15 minutes and then stained with Giemsa solution (Sigma, #G5637, USA) for 5 minutes. The numbers of single colonies containing more than 50 cells were counted under a microscope. All experiments were performed in triplicate wells, and assays were repeated three times.
Wound healing assay
The cells were seeded into 12-well plates and cultured to 100% confluence. We created a wound by scratching a straight line in the cell layer with a pipette tip. Next, the cells were washed with PBS and treated with DMEM without FBS. Cell migration was photographed and the width of the wound was measured.
Transwell migration assay
Cell culture inserts (24-well, pore size 8 µm; Sigma-Aldrich Co.) were seeded with 1×105 cells in 200 µL of medium without FBS. Medium with 5% FBS (500 µL), which served as a chemotactic agent, was added to the lower chamber. After 24 hours, non-migrating cells were removed from the upper side of the membrane, and the cells on the lower side of the membrane were fixed with 4% paraformaldehyde. Cells were stained with 0.1% crystal violet staining and counted. Each individual experiment was performed with triplicate inserts, and five microscopic fields were counted per insert.
Transwell invasion assay
Matrigel (BD Biosciences, San Jose, CA, USA) was added to each well according to the manufacturer’s instructions before the cells (3×105) were seeded on the upper chamber. Following 12 hours incubation at 37°C, non-invasive cells were gently removed from the top of the matrigel with a cotton-tipped swab. The invasive cells at the bottom of the matrigel were fixed in 4% paraformaldehyde, stained with 0.1% crystal violet, and counted. Each individual experiment was performed with triplicate inserts, and five microscopic fields were counted per insert.
Immunofluorescence assay
Anti-E-cadherin (1:200; Cell Signaling Technology), anti-N-cadherin (1:100; Abcam), and anti-SMAD4 (1:200; Abcam) were used as primary antibodies. The cells were incubated with Alexa Fluor 555/488 dye-conjugated secondary antibody (Thermo Fisher Scientific). The nucleus was stained with DAPI (Thermo Fisher Scientific).
In vivo model of HCC lung metastasis
All of the animal studies were approved by the Ruijin Hospital of Shanghai Jiaotong University. All experiments were ethically approved by the Animal Care and Use Committee of Shanghai Jiaotong University and performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.Citation18 Briefly, nude mice weighing ~20 g (Slaccas, Shanghai, China) were housed under specific pathogen-free conditions and supplied with sterilized food and water. MHCC-97H (sh-ctrl) cells were used in the metastasis assay. INTS8-knockdown cells (sh2) were suspended in PBS and 1.5×106 cells (150 µL) were injected into the tail vein of the nude mice. Mice were sacrificed 28 days after injection, and their lungs were harvested and inspected for metastatic foci. The number of metastatic nodules on the surface of H&E-stained lungs was counted under a dissecting microscope.
Statistical analysis
We used SPSS 20.0 software to perform statistical analysis. All experiments were performed at least in triplicate, and the data are presented as the means ± SDs. We assessed statistical significance using a two-tailed Student’s t-test, or, when comparing two groups, a one-way analysis of variance test with Dunnett’s test. We considered P-values of 0.05 or less as statistically significant.
Results
High INTS8 mRNA levels in HCC tissues are associated with poor overall survival
According to our analysis of the ProgeneV2 prognostic database, increased expression of INTS8 mRNA in HCC tissue is associated with poor 3-year survival rate (high INTS8, n=147; low INTS8, n=146; HR =1.47, P=0.027), and no significant difference was found in 5-year survival rate (). Moreover, according to the Kaplan–Meier Plotter database (high INTS8, n=92; low INTS8, n=272; HR =1.95, P=0.00034), higher expression of INTS8 mRNA in HCC tissue was also correlated with poor overall survival ().
Figure 1 (A) Analysis of the ProgeneV2 prognostic database: OS curve between patients with high and low integrator complex 8 (INTS8) mRNA expression. Statistical analysis was performed with the log-rank test. (B) Analysis of the Kaplan–Meier Plotter database: OS curve between patients with high and low INTS8 mRNA expression. Statistical analysis was performed with the log-rank test. (C) Immunohistochemical staining for INTS8 expression in HCC tissues. Representative image of HCC tissues and adjacent matched noncancerous tissues (magnification: 100×, 200×); *P<0.05, significantly different compared with adjacent matched noncancerous slides. (D) INTS8 levels in HCC and noncancerous tissues were assessed by WB analysis. The images are representative of five independent experiments. (E) Expression of INTS8 in human liver cancer cell lines analyzed by WB. The images are representative of five independent experiments.
Abbreviations: HCC, hepatocellular carcinoma; N, adjacent matched noncancerous tissue; OS, overall survival; T, tumor tissue; WB, Western blot.
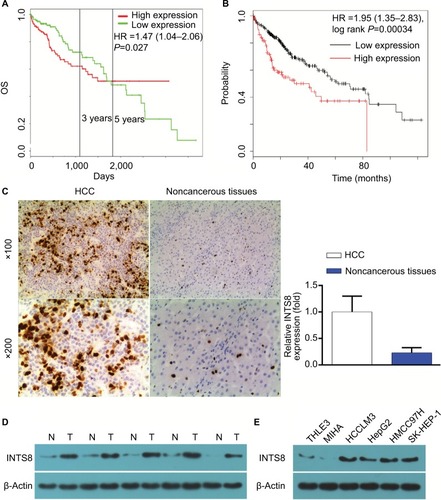
Expression of INTS8 was higher in HCC tissues than in adjacent noncancerous tissues
IHC showed that INTS8 levels were significantly higher in HCC tissues than in adjacent noncancerous tissues (; P<0.05). We examined INTS8 protein levels in HCC and adjacent noncancerous tissues by Western blotting. We observed that adjacent noncancerous tissue exhibited lower protein levels of INTS8 than HCC tissues (). Moreover, INTS8 protein levels were higher in the HCC cell lines (HCCLM3, HepG2, MHCC-97H, and SK-HEP-1) compared with the normal hepatocyte cell lines (THLE3 and MIHA) ().
Silencing of INTS8 expression in MHCC-97H and SK-HEP-1 cell lines using shRNAs
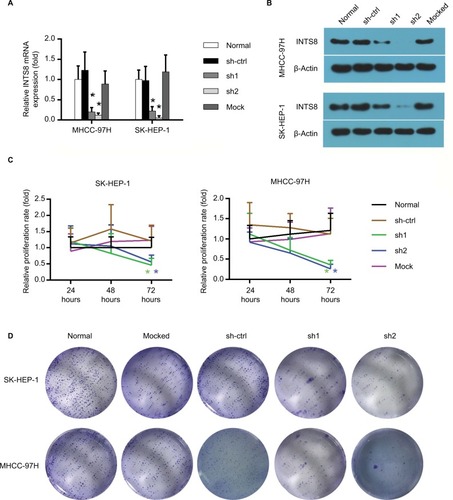
We characterized the cellular consequences of using lentiviral RNA interference vectors expressing shRNA (sh-INTS8) for stable knockdown of INTS8 expression in human HCC cell lines (MHCC-97H and SK-HEP-1) to gain insight into the possible mechanisms that mediate the relationship between INTS8 and HCC. INTS8 mRNA and protein levels were evaluated 72 hours after lentivirus infection (). Compared with the control groups, INTS8 expression was downregulated by the shRNAs. MTT analysis demonstrated that cell viability was significantly reduced as a result of INTS8 knockdown at 72 hours (). Furthermore, colony formation assay was also carried out for cell proliferation assessment. The results revealed that cell proliferation was decreased due to the downregulation of INTS8 at day 13 ().
Figure 2 Establishment of INTS8-knockdown cells and their effects on cell viability.
Notes: (A) INTS8 mRNA levels were analyzed by quantitative RT-PCR. (B) Protein expression of INTS8 was analyzed by WB. (C) Effect of INTS8 on cell viability was evaluated by MTT assay in SK-HEP-1 cells and MHCC-97H cells. The relative proliferation rate of SK-HEP-1 cells following lentivirus transfection at days 1, 2, and 3. (D) Effects of INTS8 on cell proliferation was evaluated by colony formation assay in SK-HEP-1 cells and MHCC-97H cells. The relative proliferation rate of MHCC-97H cells following lentivirus transfection at days 1, 2, and 3. The data are presented as means ± SDs. Each experiment was repeated at least five times. *P<0.05, compared with the sh-ctrl group.
Abbreviations: sh-ctrl, control short hairpin RNA group; sh1, short hairpin RNA1; sh2, short hairpin RNA2; mock, mocked treated.
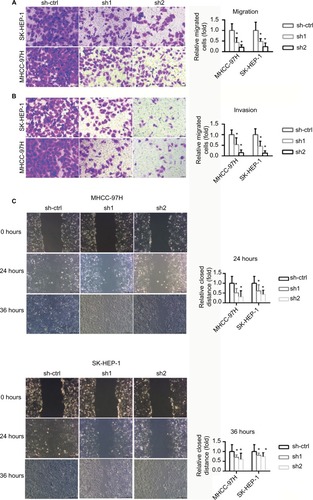
INTS8 knockdown attenuates HCC cell migration and invasion
The invasion and migratory capability of cells decreased markedly following suppression of INTS8; fewer cells migrated through the pores (). A wound-healing assay was also performed to evaluate the effect of INTS8 on HCC cell migration. INTS8 depletion significantly reduced the wound-closure capacity of HCC cell lines at 24 and 36 hours ().
Figure 3 INTS8 knockdown decreased migration and invasion by HCC cell lines.
Notes: Data are presented as means ± SDs. (A) Transwell assay was performed to assess cell migration capabilities. The number of cells was counted, with six microscopic fields counted per insert (magnification: 200×). *P <0.05, compared with the sh-ctrl group. All data are expressed as means ± SD. (B) Transwell assay was performed to assess cell invasion capabilities. The number of cells was counted, with six microscopic fields per insert (magnification: 200×). *P <0.05, compared with the sh-ctrl group. All data are expressed as means ± SD. (C) Wound-healing assay was performed to evaluate cell migration (magnification: 200×). The images are representative of five independent experiments. The relative widths of the wound gaps were measured using ImageJ software. All data are expressed as means ± SD. *P <0.05, compared with the sh-ctrl group.
Abbreviations: HCC, hepatocellular carcinoma; sh1, short hairpin RNA1; sh2, short hairpin RNA2; sh-ctrl, control short hairpin RNA group.
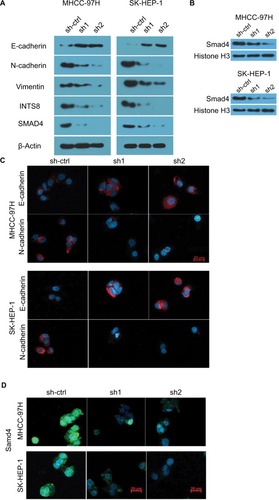
INTS8 depletion attenuates the EMT in HCC cell lines
The EMT, which plays a crucial role in cancer migration, requires alterations to molecular markers. Thus, we evaluated the expression levels of epithelial (E-cadherin) and mesenchymal (N-cadherin and vimentin) markers. WB analysis revealed that N-cadherin and vimentin were decreased by downregulation of INTS8 (). Conversely, cells with INTS8 knockdown exhibited higher expression of E-cadherin (). Immunofluorescence assessing the levels of EMT markers confirmed these results ().
Figure 4 INTS8 knockdown downregulated expression of EMT markers and Smad4.
Notes: (A) Images are representative of three independent experiments. Protein levels of E-cadherin, N-cadherin, vimentin, INTS8, and Smad4 were assessed by WB. (B) Images are representative of three independent experiments. Nuclear protein levels of Smad4 were assessed by WB. (C) Protein levels of E-cadherin and N-cadherin were assessed by immunofluorescence staining. (D) Protein levels of Smad4 were assessed by immunofluorescence staining.
Abbreviation: EMT, epithelial-to-mesenchymal transition; sh1, short hairpin RNA1; sh2, short hairpin RNA2; sh-ctrl, control short hairpin RNA group; WB, Western blot.
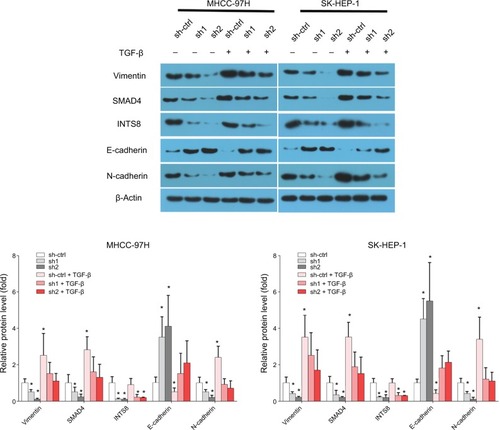
INTS8 depletion inhibits the TGF-β signaling pathway
We investigated the possible underlying molecular mechanism by evaluating the TGF-β signaling pathway. INTS8 knockdown significantly decreased the level of SMAD4, a TGF-β-activated transcription factor ().
We then investigated the effect of TGF-β pretreatment on E-cadherin, N-cadherin, and vimentin expression in INTS8-knockdown cells. According to previous studies, cells were pretreated with 5 ng/mL TGF-β1 protein for 48 hours.Citation19,Citation20 Interestingly, supplementation with TGF-β1 reduced E-cadherin levels in both mock-transfected and INTS8-knockdown cells (). Conversely, the levels of SMAD4, N-cadherin, and vimentin in both mock-transfected and INTS8-knockdown cells were significantly increased by treatment with TGF-β1 ().
Figure 5 Expression of EMT markers was rescued from INTS8 knockdown-induced suppression by TGF-β1 stimulation.
Notes: Protein levels of E-cadherin, N-cadherin, vimentin, and Smad4 were assessed by WB. The images are representative of three independent experiments. *P<0.05, compared with the sh-ctrl group.
Abbreviations: EMT, epithelial-to-mesenchymal transition; sh1, short hairpin RNA1; sh2, short hairpin RNA2; sh-ctrl, control short hairpin RNA group; WB, Western blot.
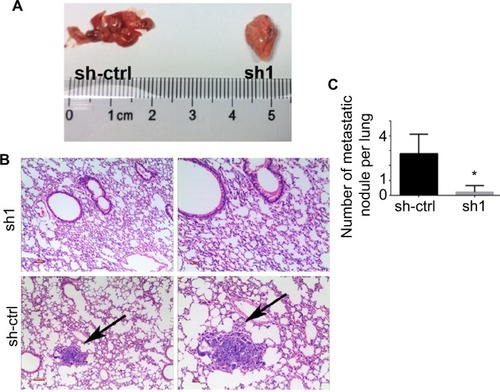
INTS8 knockdown impairs lung metastasis in vivo
We determined the role of MHCC-97H cells in HCC metastasis in vivo by injecting them into the tail vein of nude mice. As shown in , fewer metastatic nodules were detected in the lung tissues of the INTS8-knockdown group compared with the control group. Moreover, our observations of H&E-stained lung sections revealed obvious metastatic nodules in the lung tissues of the sh-ctrl group.
Figure 6 Depletion of INTS8 inhibits lung metastasis in HCC cells.
Notes: (A) Gross images of the lungs were obtained following euthanasia of the mice at 8 weeks after surgery. (B) H&E-stained lung sections. The arrows represent metastatic nodules (magnification: 100×, 200×). (C) Number of lung metastasis nodules of the INTS8-knockdown and control groups was calculated (n=5). All data are expressed as means ± SD. *P<0.05, compared with the sh-ctrl group.
Abbreviations: HCC, hepatocellular carcinoma; sh1, short hairpin RNA1; sh-ctrl, control short hairpin RNA group.
Discussion
HCC is one of the deadliest types of malignancy worldwide. Given the poor prognosis of HCC, specific molecular targets for improving the prognosis of HCC and effectiveness of intervention are urgently needed. INTS8 is a subunit of the integrator complex, which associates with the C-terminal domain of the RNA polymerase II large subunit and mediates the 3′ end processing of the small nuclear RNAs U1 and U2.Citation3 In previous studies, a significant upregulation at the protein level of INTS8 was revealed in cancer tissues and INTS8 was specifically correlated with survival.Citation4,Citation5 Interestingly, a previous study also reported that, compared with the average expression in paracancerous tissues, INTS8 levels in HCC were upregulated 2.06-fold.Citation6 Therefore, we investigated the role of INTS8 in the prognosis and progression of HCC. Our bioinformatics analysis revealed that higher levels of INTS8 expression in HCC patients were associated with poor prognoses. Next, we found that INTS8 levels were higher in HCC tissues than in adjacent noncancerous tissues. The migration and invasion of HCC cells decreased following knockdown of INTS8. Moreover, we used a model of tumor metastasis to explore its role in HCC metastasis in vivo. We detected significantly fewer metastatic nodules in the lung tissues of the mice injected with INTS8-knockdown cells, suggesting that INTS8 plays a tumor-promoting role in HCC.
The role of the EMT in the advancement of HCC is gaining increasing attention. The EMT is a complex process by which epithelial cells lose their differentiated characteristics and acquire mesenchymal features, including motility, invasiveness, and the ability to escape detection by immune cells.Citation21 The EMT is a dynamic and reversible process that allows cancer cells to reversibly transition between epithelial and mesenchymal states during metastasis.Citation22 E-cadherin is a cell–cell adhesion molecule expressed predominantly by epithelial cells. Reduction or loss of E-cadherin, which is considered a hallmark of the EMT, initiates a series of signaling events and a major reorganization of the cytoskeleton.Citation23 EMT-induced changes in epithelial plasticity are evidenced by the loss of epithelial markers, which includes the adherence junction component E-cadherin.Citation7,Citation24 Meanwhile, the expression of mesenchymal proteins such as N-cadherin and vimentin are increased.Citation7 In this study, we found that INTS8 depletion increased the expression of E-cadherin and decreased the expression of N-cadherin and vimentin. These results indicate that the expression of INTS8 is closely associated with the EMT process in HCC cells.
We have found substantial evidence that TGF-β signaling is a potent inducer of the EMT.Citation13,Citation25,Citation26 Moreover, TGF-β signaling represents a crucial pathway for initiation of the EMT in HCC.Citation2,Citation7 Yu et al reported that the effect of TGF-β on the EMT and invasion in HCC might be prevented by knockdown of myocyte enhancer factors.Citation27 TGF-β signaling is transduced through the activation of TGF-β receptors and subsequent phosphorylation of receptor-activated SMADs, which form a heterotrimeric complex with SMAD4, a transcription factor important in TGF-β signaling.Citation22,Citation28,Citation29 Interestingly, SMAD4 exerts a tumor-promoting role in HCC.Citation30 Similarly, we also found that we were able to inhibit the expression of SMAD4 by silencing the expression of INTS8. Furthermore, TGF-β1 pretreatment can be used to partly rescue the downregulated expression of SMAD4 and EMT markers induced by INTS8 depletion.
Our study had the following limitations. First, the in vitro results should be verified in other HCC cell lines. Second, our study did not reveal how INTS8 could be affecting EMT in HCC. Further studies will be needed to explore the involvement of signaling pathways. Therefore, more studies are required to demonstrate the expression of INTS8 as a potential biomarker for HCC in the clinic.
Conclusion
Taken together, our data suggest that INTS8 knockdown inhibits the TGF-β/SMAD4-induced EMT in HCC.
Author contributions
Conceived and designed the experiments: ZZ; performed the experiments: HT, XL, TL, and WQ; analyzed the data: HT, XL, TL, and WQ; contributed reagents/materials/analysis tools: ZZ; and wrote the paper: HT and ZZ. All authors contributed toward data analysis, drafting and critically revising the paper, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Acknowledgments
This study was supported by the Subject of Biological Medicine of Science and Technology Commission of Shanghai Municipality (15411950404).
Disclosure
The authors report no conflicts of interest in this work.
References
- FornerAReigMBruixJHepatocellular carcinomaLancet2018391101271301131429307467
- WangMZhangLLiuZAGO1 may influence the prognosis of hepatocellular carcinoma through TGF-β pathwayCell Death Dis20189332429487329
- BaillatDHakimiMANäärAMShilatifardACoochNShiekhattarRIntegrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase IICell2005123226527616239144
- ChengLZhangQYangSA 4-gene panel as a marker at chromosome 8q in Asian gastric cancer patientsGenomics2013102432333023722107
- SimpsonHMKhanRZSongCConcurrent mutations in ATM and genes associated with common γ chain signaling in peripheral T cell lymphomaPLoS One20151011e014190626536348
- YinFShuLLiuXMicroarray-based identification of genes associated with cancer progression and prognosis in hepatocellular carcinomaJ Exp Clin Cancer Res201635112727567667
- GiannelliGKoudelkovaPDituriFMikulitsWRole of epithelial to mesenchymal transition in hepatocellular carcinomaJ Hepatol201665479880827212245
- TanTZMiowQHMikiYEpithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patientsEMBO Mol Med20146101279129325214461
- ShrivastavaSJeengarMKThummuriDCardamonin, a chalcone, inhibits human triple negative breast cancer cell invasiveness by downregulation of Wnt/β-catenin signaling cascades and reversal of epithelial-mesenchymal transitionBiofactors201743215216927580587
- YangHDEunJWLeeKBT-cell immune regulator 1 enhances metastasis in hepatocellular carcinomaExp Mol Med2018501e42029303507
- XiaoHJiangNZhouBLiuQDuCTAZ regulates cell proliferation and epithelial-mesenchymal transition of human hepatocellular carcinomaCancer Sci2015106215115925495189
- LiangBJiaCHuangYTPX2 level correlates with hepatocellular carcinoma cell proliferation, apoptosis, and EMTDig Dis Sci20156082360237226025609
- GiannelliGVillaELahnMTransforming growth factor-β as a therapeutic target in hepatocellular carcinomaCancer Res20147471890189424638984
- LuoLMcgarveyPMadhavanSKumarRGusevYUpadhyayGDistinct lymphocyte antigens 6 (Ly6) family members Ly6D, Ly6E, Ly6K and Ly6H drive tumorigenesis and clinical outcomeOncotarget2016710111651119326862846
- GoswamiCPNakshatriHPROGgeneV2: enhancements on the existing databaseBMC Cancer20141497025518851
- LuoHHuangJLiaoWGHuangQYGaoYQThe antioxidant effects of garlic saponins protect PC12 cells from hypoxia-induced damageBr J Nutr201110581164117221205417
- LiangXLiuTZhangWZhangKGuoSLiangJLentivirus-mediated knockdown of M-phase phosphoprotein 8 inhibits proliferation of colon cancer cellsBiotechnol Appl Biochem201764691191727174009
- WorkmanPAboagyeEOBalkwillFCommittee of the National Cancer Research InstituteGuidelines for the welfare and use of animals in cancer researchBr J Cancer2010102111555157720502460
- ChenICChiangWFHuangHHChenPFShenYYChiangHCRole of SIRT1 in regulation of epithelial-to-mesenchymal transition in oral squamous cell carcinoma metastasisMol Cancer20141325425424420
- YeungTLLeungCSWongKKTGF-β modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironmentCancer Res201373165016502823824740
- JayachandranADhungelBSteelJCEpithelial-to-mesenchymal plasticity of cancer stem cells: therapeutic targets in hepatocellular carcinomaJ Hematol Oncol2016917427578206
- TangXShiLXieNSIRT7 antagonizes TGF-β signaling and inhibits breast cancer metastasisNat Commun20178131828827661
- SavagnerPEpithelial-mesenchymal transitions: from cell plasticity to concept elasticityCurr Top Dev Biol201511227330025733143
- TuWLuoMWangZUpregulation of SATB1 promotes tumor growth and metastasis in liver cancerLiver Int20123271064107822583549
- ThieryJPAcloqueHHuangRYNietoMAEpithelial-mesenchymal transitions in development and diseaseCell2009139587189019945376
- van ZijlFKrupitzaGMikulitsWInitial steps of metastasis: cell invasion and endothelial transmigrationMutat Res20117281–2233421605699
- YuWHuangCWangQMEF2 transcription factors promotes EMT and invasiveness of hepatocellular carcinoma through TGF-β1 autoregulation circuitryTumour Biol20143511109431095125087096
- HeslingCFattetLTeyreGAntagonistic regulation of EMT by TIF1γ and Smad4 in mammary epithelial cellsEMBO Rep201112766567221597466
- ZengYZhuJShenDRepression of Smad4 by miR205 moderates TGF-β-induced epithelial-mesenchymal transition in A549 cell linesInt J Oncol201649270070827279345
- HernandaPYChenKDasAMSMAD4 exerts a tumor-promoting role in hepatocellular carcinomaOncogene201534395055506825531314
