Abstract
Purpose
Increasing studies have shown that microRNA-4458 (miR-4458) is associated with human cancer progression. However, the molecular mechanism of miR-4458 in non-small-cell lung cancer (NSCLC) remains largely unknown. This study aims to reveal the biological function of miR-4458 in NSCLC.
Materials and methods
The expression of miR-4458 in NSCLC cells was evaluated by qRT-PCR. Cell proliferation and migration assay were carried out in vitro after transfection. A luciferase reporter and Western blot assay were performed to identify the functional target of miR-4458.
Results
The study indicated that miR-4458 was markedly downregulated in NSCLC cells. Overexpression of miR-4458 strongly reduced the proliferation and migration in NSCLC cell lines. In addition, miR-4458 inhibited the progression of migration and epithelial–mesenchymal transition (EMT) through the PI3K/AKT pathway. Luciferase report assay demonstrated that HMGA1 was a target gene for miR-4458.
Conclusion
The results indicate that miR-4458 participated in the process of migration and EMT via directly targeting HMGA1 and miR-4458 might be a potential novel therapeutic target in NSCLC.
The plain language summary
This study aims to reveal the biological mechanism of microRNA-4458 (miR-4458) in non-small-cell lung cancer (NSCLC). miR-4458 was markedly downregulated in NSCLC cells by qRT-PCR. Overexpression of miR-4458 strongly reduced the proliferation and migration in NSCLC cell lines. In addition, miR-4458 inhibited the progression of migration and epithelial–mesenchymal transition (EMT) through the PI3K/AKT pathway. Luciferase report assay demonstrated that HMGA1 was a target gene for miR-4458. The results indicate that miR-4458 participated in the process of migration and EMT via directly targeting HMGA1.
Introduction
Lung cancer is one of the leading causes of cancer-related deaths worldwide.Citation1 Non-small-cell lung cancer (NSCLC) accounts for ~80% of lung cancer.Citation2 Although considerable advancements have been made in diagnosis and targeted therapy for NSCLC, the prognosis is still poor.Citation3,Citation4 Therefore, it is crucial to have a better understanding of the exact mechanism for the development and progression of NSCLC, which could provide more individualized and effective therapeutic strategies for NSCLC patients.
miRNAs are a class of small (22–24 nucleotides) ncRNAs that play a pivotal role in the diagnosis and prognosis of malignant neoplasm.Citation5–Citation7 Previous studies have already found that miRNAs could inhibit the transcription of target mRNAs via binding to complementary 3′-UTR. Emerging evidence indicates that microRNA-4458 (miR-4458) plays an important role in different cell processes, including proliferation, cell cycle, and glycolysis in hepatocellular carcinoma,Citation8 colon cancer,Citation9 and lung cancer.Citation10,Citation11 However, the molecular mechanism of miR-4458 in NSCLC has not been fully understood. Therefore, the understanding of the biological activities utilized by miR-4458 in NSCLC is urgently required.
The HMGA1 serves as a regulator of the chromatin structure via direct binding to A/T-rich DNA sequences.Citation12 Studies find that HMGA1 plays a carcinogenic role in various cancer types, such as thyroid cancer,Citation13 breast cancer,Citation14 and lung cancer.Citation15 Accumulating evidence shows that HMGA1 is associated with biological processes of cell proliferation, cell cycle, and metastasis.Citation16,Citation17 Moreover, overexpression of HMGA1 leads to the promotion of epithelial–mesenchymal transition (EMT) in basal-like breast cancer.Citation18 In addition, HMGA1 could be regulated by miRNAs, such as miR-26aCitation19 and miR-625.Citation20 However, its role and the molecular mechanism in NSCLC still remain obscure.
In the present study, we demonstrated that miR-4458 inhibited proliferation and migration in NSCLC cells. It was shown that miR-4458 suppressed the progression of migration and EMT through the PI3K/AKT pathway. Furthermore, we explored and validated HMGA1 as a direct target of miR-4458. Thus, our results suggest that miR-4458 might be a potential therapeutic target in NSCLC.
Materials and methods
Cell culture and transfection
A549, H1299, HCC827, PC9, HBE, 293 T cell lines were purchased from American Type Culture Collection (Manas-sas, VA, USA). All cells were cultured at 37°C in an incubator with 5% CO2. miR-4458 mimics (mimics), negative control (NC), miR-4458 inhibitor (inhibitor), and inhibitor negative control (inhibitor NC) were used (RiboBio, Guangzhou, China) for the overexpression and knockdown of miR-4458. The si-HMGA1 was conducted for the knockdown of HMGA1 and si-NC was used as the control. Transfection of cells was performed using riboFECT™ CP Transfection Agent (RiboBio, Guangzhou, China) with a 100 nM concentration following the manufacturer’s protocol.
qRT-PCR analysis
The total RNA was extracted from cultured cells using TRIzol (Thermo Fisher Scientific, Waltham, MA, USA). The expression level of miR-4458 was assessed by Hairpin-it™ microRNA RT-PCR Quantitation Kit (Genepharma, Shanghai, China) while the mRNA expression was measured by a SYBR Premix Ex Taq II kit (TaKaRa, Dalian, China). β-actin and U6 were used as an internal control. miR-4458 and mRNA expression was evaluated using Light Cycler 480 System II (Roche).
EdU assay
After 48 hours of transfection, 5-ethynyl-2′-deoxyuridine (Edu) was added into A549 and H1299 cells with 2-hour incubation at 37°C. Then, the cell proliferation assay was performed by Cell-Light EdU Apollo®567 In Vitro Imaging Kit (RiboBio, Guangzhou, China) according to the manufacturer’s protocol.
Cell counting kit-8 (CCK-8) assay
Cells were seeded into 96-well plates (2×103 cells per well) after 24 hours of transfection. Cell proliferation assay was assessed by CCK-8 assay (Dojindo, Kumamoto, Japan) using a microplate reader (Thermo Fisher Scientific) at 450 nm wavelength. The assay was repeated six times at each time point.
Colony formation assay
For colony formation assay, A549 and H1299 cells transfected with miR-4458 mimics and inhibitors were seeded into 6-well plates with a density of 200 cells per well. After culturing in the cell incubator (37°C, 5% CO2) for 14 days, the cells were fixed in 70% ethanol and stained by crystal violet.
Cell migration assay
In order to detect the migratory capability, 2×105 cells were seeded into a transwell apparatus (Corning, NY, USA) with the serum-free medium. Relatively, medium containing 10% serum functioned as a chemoattractant in the lower chamber. After incubation for 12 hours, the stained cells were photographed and counted by a microscope.
Wound healing assay
A549 and H1299 cells transfected with miR-4458 mimics, inhibitors, and si-HMGA1 were seeded into six-well plates to perform wound healing assay. When the fusion of cells reached above 90%, the scratch was marked by using a standard 10 µL pipette tip at the middle of the well. Cells were further cultured with serum-free medium for 24 hours. The inverted microscope was used to observe the healing of the wound.
Western blotting analysis
Cells were harvested and lysed by RIPA buffer (Beyotime, Shanghai, China) after 48 hours of transfection. After the separation of protein using 10% SDS-PAGE and the transfer using polyvinylidene fluoride membrane, the antibodies were incubated at 4°C overnight after blocking for 2 hours with 5% skim milk. The antibodies against HMGA1, E-cadherin, N-cadherin, Snail, AKT, and p-AKT were obtained from Abcam (Cambridge, UK) while β-actin antibody served as a loading control. An ECL Advanced Western Blot Detection Kit (Thermo Fisher Scientific) was used to visualize the specific protein bands. The Image Lab software was used for analyzing the results from Western blot. The experiments were repeated three times.
Bioinformatics analysis
Publicly available databases were used to predict potential targets of miR-4458, including TargetScan (www.targetscan.org/), TarBase (http://diana.imis.athena-innovation.gr/DianaTDian/index.php?r=tarbase/index), and miRwalk (http://mirwalk.umm.uni-heidelberg.de/). We explored common target genes from three databases and then carried out the analysis of Gene Ontology (GO) and pathway enrichment using Funrich software. In order to explore more possible targets, the expression profile data of lung adenocarcinoma (LUAD) from the The Cancer Genome Atlas (TCGA) database including 73 cancer samples and 59 normal samples were analyzed. GSE32863 and GSE10072 datasets were explored to define the expression of HMGA1 between LUAD and normal group by Oncomine database (www.oncomine.org/resource/login.html). For the understanding of prognostic information, the free available Kaplan–Meier plotter database (www.kmplot.com) was used to analyze the overall survival (OS) of HMGA1. Cancer Cell Line Encyclopedia (CCLE) database (https://portals.broadinstitute.org/ccle) was used to visualize the expression of HMGA1 in lung cancer cell lines.
Dual luciferase reporter assay
First, cells (1.5×104) were plated on 96-well plates. Next, cells were cotransfected with different combinations of miR-4458, miR-NC, h-HMGA1-wild type (WT), and h-HMGA1-mutant type (MUT) after 24 hours. Finally, the relative luciferase signals were assessed after 48 hours using the Dual Luciferase Reporter Assay Kit (Promega Corporation, Fitchburg, WI, USA).
Statistical analyses
All statistical analysis was carried out using GraphPad Prism 6.0 software (La Jolla, CA, USA). Results were expressed as the mean ± SD among triplicate samples. One-way ANOVA or two-tailed Student’s t-test was applied to evaluate statistical differences. P<0.05 was considered to be statistically significant.
Results
miR-4458 inhibited cell proliferation and colony formation in NSCLC cells
qRT-PCR was employed to investigate the expression of miR-4458. As expected, miR-4458 was significantly down-regulated in four NSCLC cell lines (A549, H1299, HCC827, and PC9) compared with one normal lung cell (HBE, ). Higher expression of miR-4458 was discovered in NSCLC cells transfected with miR-4458 mimics. Meanwhile, the expression of miR-4458 was lower in NSCLC cells transfected with miR-4458 inhibitor (). In order to monitor whether miR-4458 could influence the proliferation in NSCLC cells, we performed Edu and CCK-8 assay. The results of Edu assay indicated that overexpression of miR-4458 inhibited proliferation. In contrast, downregulation of miR-4458 enhanced cell growth of NSCLC cells (). Furthermore, CCK-8 assay confirmed that miR-4458 overexpression suppressed the proliferation rate, whereas miR-4458 knockdown increased growth of NSCLC cells compared with the NC group (). Consistent with these findings, colony formation ability was reduced with the overexpression of miR-4458 and enhanced with the knockdown of miR-4458 (). In conclusion, these results demonstrated that miR-4458 inhibited cell proliferation and colony formation in NSCLC cells.
Figure 1 miR-4458 inhibited cell proliferation in NSCLC cells.
Notes: (A) Relative expression level of miR-4458 in four NSCLC cell lines (A549, H1299, HCC827, and PC9) and one normal lung cell (HBE). (B) The expression of miR-4458 after 48 hours of transfection with NC, mimics, inhibitor NC, and inhibitor by qRT-PCR. (C) The rate of cell growth was measured by Edu assay. (D) CCK-8 assay confirmed that miR-4458 inhibited cell proliferation in NSCLC cells. (E) The ability of colony formation was reduced with the overexpression of miR-4458 and enhanced with knockdown of miR-4458. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: CCK-8, cell-counting kit-8; Edu, 5-ethynyl-2′-deoxyuridine; miR-4458, microRNA-4458; NC, negative control; NSCLC, non-small-cell lung cancer.
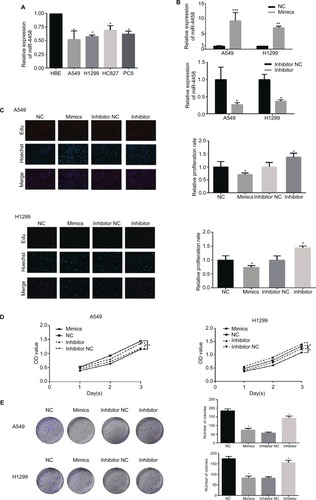
miR-4458 inhibited migration and the EMT process in NSCLC cells
In order to explore the migratory capability of miR-4458 in NSCLC, we carried out transwell migration assay. The results showed that miR-4458 overexpression significantly inhibited migration in NSCLC cells. Moreover, knockdown of miR-4458 led to an increase in migration compared with the NC group (). Furthermore, wound healing assay confirmed that miR-4458 overexpression suppressed migration, whereas miR-4458 knockdown increased migration of NSCLC cells compared with the NC group (). To determine the effect of miR-4458 on EMT, we performed Western blotting to evaluate the expression of EMT biomarkers (E-cadherin, N-cadherin, and Snail) after 48 hours of transfection. The findings revealed that miR-4458 significantly upregulated the expression of E-cadherin and downregulated the level of N-cadherin and Snail. With knockdown of miR-4458, E-cadherin was reduced while N-cadherin and Snail were increased (). These results suggested that miR-4458 played a pivotal role in the process of migration and EMT.
Figure 2 miR-4458 inhibited migration and the EMT process in NSCLC cells
Notes: (A) The results of transwell assay showed that miR-4458 reduced migration ability in NSCLC cells. (B) The results of wound-healing assay showed that miR-4458 inhibited migration ability. (C) Western blot analysis indicated that overexpression of miR-4458 upregulated EMT biomarker E-cadherin and reduced EMT biomarkers N-cadherin and Snail in NSCLC cells. Relatively, E-cadherin was downregulated and N-cadherin and Snail were enhanced with miR-4458 knockdown. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: EMT, epithelial–mesenchymal transition; miR-4458, microRNA-4458; NC, negative control; NSCLC, non-small-cell lung cancer.
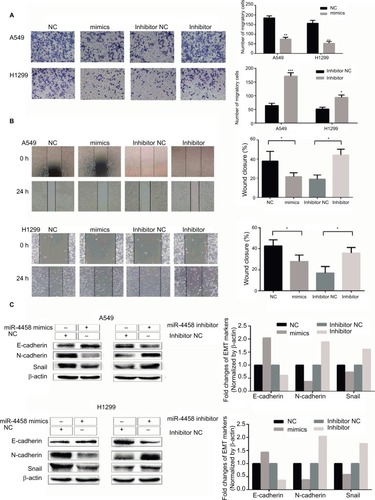
miR-4458 inhibited migration and the EMT process via PI3K/AKT pathway in NSCLC cells
PI3K/AKT pathway plays a key role in promoting migration and the EMT process in NSCLC cells. To investigate whether miR-4458 could reduce migration and the EMT process through PI3K/AKT signaling pathway, Western blot assay was carried out. The results indicated that overexpression of miR-4458 obviously reduced the level of p-AKT in NSCLC cells. Consistently, knockdown of miR-4458 led to a robust increase in the expression of p-AKT compared with control group (). In addition, we cultured NSCLC cells with a PI3K inhibitor (LY294002). The results of transwell assay demonstrated that cells treated with LY294002 inhibited migration compared with control group (). Moreover, our results showed that LY294002 group enhanced the expression of E-cadherin and reduced the level of N-cadherin, Snail, and p-AKT (). These findings showed that miR-4458 was involved in the migration and the EMT process through PI3K/AKT pathway in NSCLC.
Figure 3 miR-4458 inhibited migration and the EMT process via PI3K/AKT pathway in NSCLC cells.
Notes: (A) The protein levels of p-AKT and AKT were evaluated by Western blot assay. (B) The results of transwell assay with miR-4458 knockdown were abolished by a PI3K inhibitor (LY294002). (C) The protein levels of E-cadherin, N-cadherin, Snail, p-AKT, and AKT were evaluated by the exposure of LY294002. **P<0.01.
Abbreviations: EMT, epithelial–mesenchymal transition; miR-4458, microRNA-4458; NSCLC, non-small-cell lung cancer.
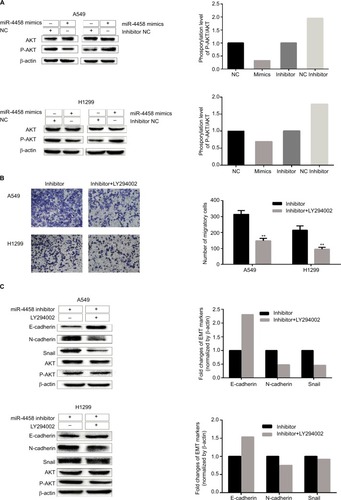
Bioinformatics analysis of the targets of miR-4458
Three publicly available databases (TargetScan, TarBase, and miRwalk) were explored to define the targets of miR-4458 (). One hundred seventy-six common target genes were used for the analysis of GO and pathway enrichment (). After the exploration of LUAD expression profile data from the TCGA database, eight more possible targets (HMGA1, VANGL2, E2F2, STRBP, RRP1B, FAXC, SYT7, and CASK) were screened out (). Notably, we selected HMGA1 to do further research. First, the expression of HMGA1 was upregulated in LUAD group compared with normal group in GSE32863 and GSE10072 datasets (). Second, higher HMGA1 expression was correlated with poor progression of NSCLC from Kaplan–Meier plotter database (). Finally, CCLE database indicated that HMGA1 was highly expressed in lung cancer cell lines (). The qRT-PCR assay confirmed that HMGA1 was upregulated in four NSCLC cell lines (A549, H1299, HCC827, and PC9) compared with HBE ().
Figure 4 Bioinformatics analysis of the targets of miR-4458.
Notes: (A) Venn chart of common target genes from three publicly available databases including TargetScan, TarBase, and miRwalk. (B) GO analysis included biological processes, cellular component, molecular function, and pathway enrichment analysis for 176 common targets. (C) Venn chart of 8 more possible targets from 176 common target genes and TCGA database. (D) The expression of HMGA1 between LUAD and normal group in GSE32863 and GSE10072 datasets. (E) Analysis of the OS about HMGA1 from Kaplan–Meier plotter database. (F) The expression of HMGA1 in lung cancer cell lines from CCLE database. (G) The expression of HMGA1 between four NSCLC cell lines (A549, H1299, HCC827, and PC9) and one normal lung cell (HBE) by qRT-PCR. *P<0.05, **P<0.01.
Abbreviations: GO, Gene Ontology; miR-4458, microRNA-4458; NC, negative control; OS, overall survival.
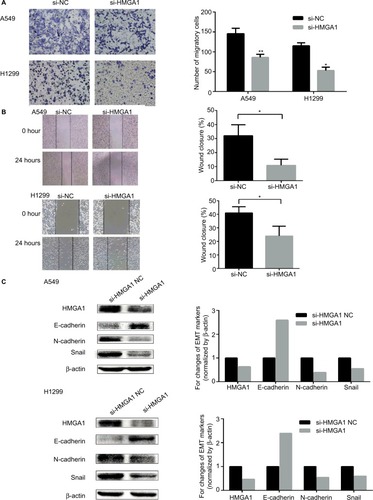
HMGA1 enhanced migration and the EMT process in NSCLC cells
In order to confirm the function of HMGA1 in the migration and EMT process, transwell assay, wound healing assay, and Western blot assay were carried out in NSCLC cells. As shown in , cells transfected with si-HMGA1 showed a significant reduction in migration compared with NC group. The results of wound healing assay indicated that HMGA1 knockdown inhibited migration in NSCLC cell lines (). The expression of EMT biomarker E-cadherin was increased and EMT biomarker N-cadherin and Snail were reduced after HMGA1 was knocked down (). These findings demonstrated that HMGA1 played an important role in regulating metastasis and EMT process in NSCLC cells.
Figure 5 HMGA1 enhanced migration and the EMT process in NSCLC cells.
Notes: (A) The results of transwell assay showed that downregulation of HMGA1 reduced migration capacity in NSCLC cells. (B) The results of wound-healing assay showed that downregulation of HMGA1 inhibited migration capacity in NSCLC cells. (C) Western blot assay indicated that EMT biomarker E-cadherin was upregulated and N-cadherin and Snail were downregulated after knockdown of HMGA1. *P<0.05, **P<0.01.
Abbreviations: EMT, epithelial–mesenchymal transition; miR-4458, microRNA-4458; NC, negative control; NSCLC, non-small-cell lung cancer.
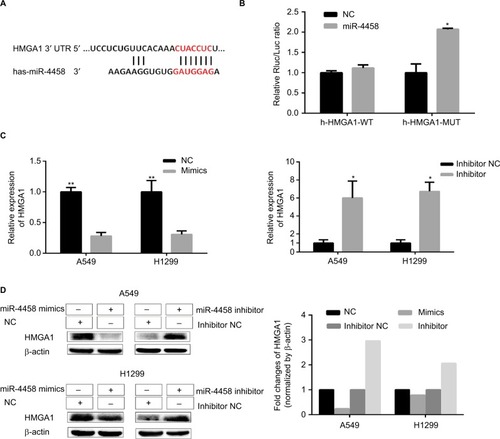
HMGA1 was a direct target of miR-4458
The predicted binding site of miR-4458 in the 3′-UTR of HMGA1 was obtained from miRNA website (). The results revealed that luciferase activity cotransfected with combinations of miR-4458 and h-HMGA1-MUT was increased compared with combinations of miR-4458 and h-HMGA1-WT (). To confirm the expression of HMGA1 in NSCLC cells, qRT-PCR and Western blot were conducted after 48 hours of transfection with miR-4458 NC, mimics, inhibitor NC, and inhibitor. Overexpression of miR-4458 obviously inhibited both mRNA and protein expression of HMGA1. Contrastively, miR-4458 knockdown led to a sharp increase of HMGA1 ().
Figure 6 HMGA1 was a direct target of miR-4458.
Notes: (A) The predicted binding site of miR-4458 in the 3′-UTR of HMGA1 from miRNA website. (B) The analysis of luciferase activity after different combinations of miR-4458, miR-NC, h-HMGA1-WT, and h-HMGA1-MUT. (C) The expression of HMGA1 in NSCLC cells after 48 hours of transfection with miR-4458 NC, mimics, inhibitor NC, and inhibitor. (D) Western blot analysis of HMGA1 in NSCLC cells after 48 hours of transfection with miR-4458 NC, mimics, inhibitor NC, and inhibitor. *P<0.05, **P<0.01.
Abbreviations: miR-4458, microRNA-4458; NC, negative control; NSCLC, non-small-cell lung cancer.
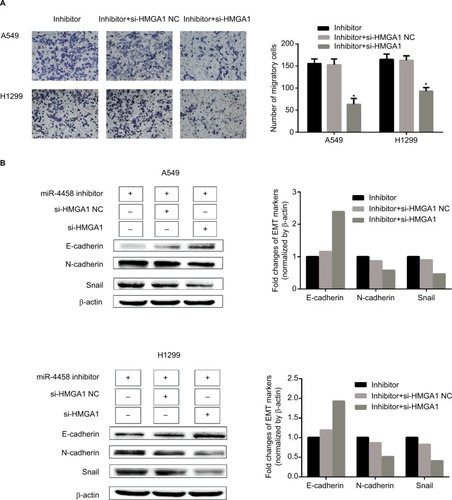
Knockdown of HMGA1 abolished the role of downregulated miR-4458 in the process of migration and EMT
The rescue experiment was performed to demonstrate whether HMGA1 could serve as a controller of miR-4458 in the process of migration and EMT. Here are three groups, which were cells transfected with miR-4458 inhibitor, cells cotransfected with miR-4458 inhibitor and si-HMGA1 NC, and cells cotransfected with miR-4458 inhibitor and si-HMGA1. Transwell assay was conducted after the transfection. Compared with miR-4458 inhibitor transfected cells, the migratory capability of miR-4458 significantly decreased in cells cotransfected with miR-4458 inhibitor and si-HMGA1 (). The results of Western blot assay showed that the expression of E-cadherin increased in cells cotransfected with miR-4458 inhibitor and si-HMGA1. Moreover, the expression of N-cadherin and Snail reduced under the same comparison (). Taken together, these findings showed that miR-4458 inhibited migration and EMT process depending on the regulation of HMGA1 in NSCLC.
Figure 7 Knockdown of HMGA1 abolished the role of downregulated miR-4458 in the process of migration and EMT.
Notes: (A) A549 and H1299 were cotransfected with miR-4458 inhibitor, si-HMGA1 NC and si-HMGA1, and the transwell results were shown after 48 hours of transfection. (B) Western blot analysis was carried out after 48 hours of transfection.
Abbreviations: EMT, epithelial–mesenchymal transition; miR-4458, microRNA-4458; NC, negative control.
Discussion
It is well known that miRNAs play an important role in the diagnosis and prognosis via binding to 3′-UTR of targets in various malignant neoplasm.Citation5,Citation6 In spite of the increasing amount of miRNAs in the development of tumorigenesis, the specific functional molecular mechanism remains unclear. miR-4458 is first detected to be a tumor suppressor in hepatocellular carcinoma.Citation8 Moreover, overexpression of miR-4458 suppresses glycolysis and lactate production via targeting hexokinase2 in colon cancer.Citation9 Emerging evidence indicates that miR-4458 inhibits proliferation by directly targeting CCND1 and Lin28B in lung cancer,Citation10,Citation11 which is in accordance with the results of cell growth in our study. In the present research, we confirmed that miR-4458 inhibited cell proliferation () and colony formation ability in vitro (). In addition, the expression of the miR-4458 was downregulated in NSCLC cell lines by qRT-PCR (). Thus, the results suggest that miR-4458 might inhibit proliferation and colony formation in NSCLC cells.
EMT serves as an important event in the tumor metastasis progression.Citation21–Citation23 In the present study, for the first time, we found that miR-4458 suppressed migration and EMT bio-markers in NSCLC cells. First, we performed transwell assay and wound-healing assay to make sure the role of miR-4458 in the development of migration in NSCLC cell. The results showed that miR-4458 knockdown enhanced the number of migratory cells and the width of scratch compared with NC group (). Notably, we further conducted the experiment in the analysis of EMT process. Downregulation of E-cadherin and overexpression of N-cadherin and Snail are considered as key elements in EMT process.Citation24,Citation25 The results discovered that miR-4458 influenced the expression of EMT biomarkers by Western blotting. Overexpression of miR-4458 significantly increased the expression of E-cadherin and reduced the level of N-cadherin and Snail. In contrast, E-cadherin was reduced while N-cadherin and Snail were increased with miR-4458 knockdown (). The results clearly indicate that miR-4458 negatively mediates cell migration and process of EMT through regulating E-cadherin, N-cadherin, and Snail. PI3K/AKT is a pivotal signal pathway in malignant tumor progression and is associated with cell proliferation, migration, and EMT process.Citation26–Citation29 In the present study, further studies were carried out to confirm the role of PI3K/AKT pathway in the molecular mechanism of miR-4458. First, the results of Western blot indicated that the level of p-AKT was reduced in NSCLC cells with overexpression of miR-4458. Meanwhile, miR-4458 knockdown enhanced the expression of p-AKT compared with control group (). Second, NSCLC cells were cultured with a PI3K inhibitor (LY294002) in order to explore the further mechanism. The results of transwell assay with miR-4458 knockdown were abolished by LY294002 (). Moreover, our results showed that LY294002 group enhanced the expression of E-cadherin and reduced the level of N-cadherin, Snail, and p-AKT (). Notably, the results of bioinformatics analysis with 176 common targets suggested that PI3K/AKT activation might be involved in the biological pathway of miR-4458 (). These findings show that miR-4458 might play an important role in the migration and EMT process through the PI3K/AKT pathway in NSCLC.
Recently, a large number of bioinformatics analysis methods have been used to explore the molecular mechanism of laryngeal cancer with the advantage of promoting credibility and reliability.Citation30–Citation32 For the analysis of bioinformatics methods in our study, we screened out 176 common target genes from three databases (TargetScan, TarBase, and miRwalk, ). Then, we carried out GO (biological processes, cellular component, molecular function) and pathway enrichment analysis by Funrich software (). Additionally, the LUAD database from TCGA was explored to confirm the more possible targets. Finally, eight targets (HMGA1, VANGL2, E2F2, STRBP, RRP1B, FAXC, SYT7, and CASK) were discovered (). Notably, we selected HMGA1 to do further bioinformatics analysis. The results of Oncomine database suggested that the expression of HMGA1 in LUAD tissues was significantly upregulated compared with normal group from two Gene Expression Omnibus datasets (). Higher HMGA1 expression was correlated with poor progression of both lung adenocarcinoma and lung squamous cell carcinoma from Kaplan–Meier plotter database (). The level of HMGA1 in lung cancer cells was obtained from CCLE database (). These bioinformatics analysis results indicate that HMGA1 may be involved in the tumorigenesis and progression of NSCLC as a potential target of miR-4458. HMGA1 functions as a regulator of the chromatin structure via direct binding to A/T-rich DNA sequences.Citation12 Accumulating evidence indicates that HMGA1 plays a carcinogenic role in various cancer process, including cell proliferation, cell cycle, and metastasis.Citation13–Citation16 In NSCLC, HMGA1 is correlated with the malignant status and prognosis.Citation17 Additionally, HMGA1 leads to the promotion of EMT in basal-like breast cancer.Citation18 In our study, we explored the function of HMGA1 in the process of migration and EMT. For transwell assay, knockdown of HMGA1 reduced migration capacity in NSCLC cells (). Similarly, wound-healing assay showed that knockdown of HMGA1 inhibited migration capacity (). Western blot assay indicated that EMT biomarker E-cadherin was upregulated and N-cadherin and Snail were downregulated after knockdown of HMGA1 ().
It has been demonstrated that multiple miRNAs could target the same gene. In lung adenocarcinoma cells, miR-26a had a negative correlation with HMGA1.Citation19 In breast cancer, miR-625 suppressed cell growth and migration via targeting HMGA1.Citation20 However, it is the first time to report that HMGA1 is a direct target of miR-4458 in NSCLC. In our study, we revealed the regulatory relationship of miR-4458 and HMGA1 in NSCLC cells. The luciferase assay confirmed that miR-4458 was directly binding to the 3′-UTR of HMGA1 (). In order to confirm the expression of both mRNA and protein in NSCLC cells, we carried out qRT-PCR and Western blot assay to underpinned the results (). Mechanistically, the rescue assay revealed that knockdown of HMGA1 abolished the role of downregulated miR-4458 in the migration and EMT process ().
Conclusion
In summary, our study demonstrated that miR-4458 inhibited proliferation and migration in NSCLC cells. In addition, miR-4458 significantly suppressed the progression of metastasis and EMT via the PI3K/AKT pathway. Moreover, miR-4458 participated in the process of migration and EMT process via directly binding to the 3′-UTR of HMGA1. Thus, our results suggest that miR-4458 might be a potential therapeutic target for NSCLC treatment.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 81771867, No. 81771868).
Disclosure
The authors report no conflicts of interest in this work.
References
- TorreLABrayFSiegelRLFerlayJLortet-TieulentJJemalAGlobal cancer statistics, 2012CA Cancer J Clin20156528710825651787
- Longo-SorbelloGSChenBBudak-AlpdoganTBertinoJRRole of pemetrexed in non-small cell lung cancerCancer Invest2007251596617364559
- WoodDEKazerooniEABaumSLLung cancer screening, version 3.2018, NCCN clinical practice guidelines in oncologyJ Natl Compr Canc Netw201816441244129632061
- SaintignyPBurgerJARecent advances in non-small cell lung cancer biology and clinical managementDiscov Med2012137128729722541616
- BartelDPMicroRNAs: genomics, biogenesis, mechanism, and functionCell2004116228129714744438
- ChenCZMicroRNAs as oncogenes and tumor suppressorsN Engl J Med2005353171768177116251533
- KangWHuangTZhouYmiR-375 is involved in Hippo pathway by targeting YAP1/TEAD4-CTGF axis in gastric carcinogenesisCell Death Dis2018929229367737
- TangDSunBYuHYangZZhuLTumor-suppressing effect of miR-4458 on human hepatocellular carcinomaCell Physiol Biochem20153551797180725833000
- QinYChengCLuHWangYmiR-4458 suppresses glycolysis and lactate production by directly targeting hexokinase2 in colon cancer cellsBiochem Biophys Res Commun20164691374326607110
- BaoLWangLWeiGWangYWuyunGBoARole of microRNA-4458 in patients with non-small-cell lung cancerOncol Lett20161253958396627895756
- LiuCHLvDSLiMSunGZhangXFBaiYMicroRNA-4458 suppresses the proliferation of human lung cancer cells in vitro by directly targeting Lin28BActa Pharmacol Sin20173891297130428603287
- FuscoAFedeleMRoles of HMGA proteins in cancerNat Rev Cancer200771289991018004397
- ChiappettaGTalliniGde BiasioMCDetection of high mobility group I HMGI(Y) protein in the diagnosis of thyroid tumors: HMGI(Y) expression represents a potential diagnostic indicator of carcinomaCancer Res19985818419341989751634
- HuangRHuangDDaiWYangFOverexpression of HMGA1 correlates with the malignant status and prognosis of breast cancerMol Cell Biochem20154041–225125725772486
- LinSYPengFAssociation of SIRT1 and HMGA1 expression in non-small cell lung cancerOncol Lett201611178278826834854
- ZhongJLiuCZhangQHTGF-β1 induces HMGA1 expression: the role of HMGA1 in thyroid cancer proliferation and invasionInt J Oncol20175051567157828393241
- ZhangZWangQChenFLiuJElevated expression of HMGA1 correlates with the malignant status and prognosis of non-small cell lung cancerTumour Biol20153621213121925344216
- PegoraroSRosGPiazzaSHMGA1 promotes metastatic processes in basal-like breast cancer regulating EMT and stemnessOncotarget2013481293130823945276
- SekimotoNSuzukiASuzukiYSuganoSExpression of miR-26a exhibits a negative correlation with HMGA1 and regulates cancer progression by targeting HMGA1 in lung adenocarcinoma cellsMol Med Rep201715253454228000891
- ZhouWBZhongCNLuoXPmiR-625 suppresses cell proliferation and migration by targeting HMGA1 in breast cancerBiochem Biophys Res Commun2016470483884426806308
- LarueLBellacosaAEpithelial–mesenchymal transition in development and cancer: role of phosphatidylinositol 3′ kinase/AKT pathwaysOncogene200524507443745416288291
- YilmazMChristoforiGEMT, the cytoskeleton, and cancer cell invasionCancer Metastasis Rev2009281–2153319169796
- QureshiRAroraHRizviMAEMT in cervical cancer: its role in tumour progression and response to therapyCancer Lett2015356232133125281477
- HendersonVSmithBBurtonLJRandleDMorrisMOdero-MarahVASnail promotes cell migration through PI3K/AKT-dependent Rac1 activation as well as PI3K/AKT-independent pathways during prostate cancer progressionCell Adh Migr20159425526426207671
- HuberMAKrautNBeugHMolecular requirements for epithelial– mesenchymal transition during tumor progressionCurr Opin Cell Biol200517554855816098727
- Fresno VaraJACasadoEde CastroJCejasPBelda-IniestaCGonzález-BarónMPI3K/Akt signalling pathway and cancerCancer Treat Rev200430219320415023437
- FuQFLiuYFanYAlpha-enolase promotes cell glycolysis, growth, migration, and invasion in non-small cell lung cancer through FAK-mediated PI3K/AKT pathwayJ Hematol Oncol201582225887760
- DanielsenSAEidePWNesbakkenAGurenTLeitheELotheRAPortrait of the PI3K/AKT pathway in colorectal cancerBiochim Biophys Acta20151855110412125450577
- XuWYangZLuNA new role for the PI3K/Akt signaling pathway in the epithelial–mesenchymal transitionCell Adh Migr20159431732426241004
- ZhangHWSunXFHeYNLiJTGuoXHLiuHBioinformatics analysis of breast cancer bone metastasis related gene-CXCR4Asian Pac J Trop Med20136973273823827153
- VishnubalajiRHamamRAbdullaMHGenome-wide mRNA and miRNA expression profiling reveal multiple regulatory networks in colorectal cancerCell Death Dis201561e161425611389
- Emmert-StreibFde Matos SimoesRGlazkoGFunctional and genetic analysis of the colon cancer networkBMC Bioinformatics201415Suppl 6S6
