Abstract
Background
Although the number of circular RNAs (circRNAs) that has been identified in multiple cancer tissues continues to increase, the relationship between circRNA expression and carcinogenesis remains unknown. The role of hsa_circ_0072309 in breast cancer has remained undefined until now. In this study, we aimed to investigate the role of hsa_circ_0072309 in breast cancer progression.
Methods
hsa_circ_0072309 expression in breast cancer tissues was analyzed using qRT-PCR. A series of functional experiments were carried out to investigate hsa_circ_0072309 function in breast cancer development and its underlying molecular mechanisms.
Results
hsa_circ_0072309 expression in breast cancer tissues was upregulated relative to that in adjacent normal tissues. hsa_circ_0072309 could serve as a prognostic biomarker of breast cancer. hsa_circ_0072309 overexpression dramatically inhibited the proliferation, migration, and invasion of breast cancer cells in vitro. In vivo assays revealed that the ectopic expression of hsa_circ_0072309 repressed breast cancer growth. The results of our mechanistic studies indicated that hsa_circ_0072309 could act as the sponge of miR-492, which exhibited increased expression in breast cancer tissues. Hsa_circ_0072309 suppressed breast cancer cell proliferation, migration, and invasion by inhibiting miR-492.
Conclusion
Our findings revealed for the first time that the hsa_circ_0072309-miR-492 axis plays an essential role in breast cancer progression.
Introduction
The incidence of breast cancer, the solid tumor class that causes the highest death rate among women worldwide,Citation1,Citation2 continues to increase. This malignancy will affect 12% of all women, and approximately 30%–40% of patients with breast cancer will die.Citation3 Numerous therapies for breast cancer have been developed. These therapies, however, have demonstrated limited efficacy. Adjuvant therapy and hormonal treatment improve the survival rates and prognosis of patients with breast cancer but exhibit severe side effects that restrict their usage.Citation3 Thus, methods for the early diagnosis, accurate prediction, and appropriate treatment of breast cancer still need to be developed. Understanding the molecular pathways involved in the pathogenesis of breast cancer is necessary to maximize treatment efficiency.Citation4
Circular RNAs (circRNAs) are a group of non-coding RNAs that lack 5′ caps or 3′ poly-A tails.Citation5 These non-coding RNAs were originally considered as splicing errors.Citation6 Nevertheless, evidence showing that circRNAs are not simply the by-products of splicing errors and may regulate cellular function through distinct mechanisms continues to accu mulate.Citation7 Many circRNAs are dysregulated in cancer progression.Citation8,Citation9 Mechanistic studies have shown that circRNAs sponge miRNA expression and regulate biological processes.Citation10,Citation11 For example, circRNA ciRS-7 functions as an miRNA inhibitor by sponging miR-7.Citation12 Information on the function of circRNAs in breast cancer progression, however, remains limited.
In this study, we investigated the function of hsa_ circ_0072309 in breast cancer progression. By using a public dataset (GSE77661), we found that circ_0072309 expression in breast cancer tissues was lower than that in normal control tissues. Thus, we selected this circRNA for further investigation. We discovered that hsa_circ_0072309 was downregulated in breast cancer cells and tumor tissues and that the downregulation of hsa_circ_0072309 was associated with the poor survival rate of patients with breast cancer. Overexpression experiments showed that hsa_circ_0072309 inhibited breast cancer progression in vitro and in vivo by regulating the proliferation of breast cancer cells. The results of our mechanistic studies revealed that hsa_circ_0072309 sponged miR-492. This effect consequently inhibited miR-492 and thus suppressed tumor progression. Our findings provide evidence that hsa_circ_0072309 plays an important role in breast cancer progression.
Materials and methods
Patient specimens
A total of 32 breast cancer tissues and matched normal tissues were obtained from The First Affiliated Hospital of Wenzhou Medical University. All tissue samples were stored in liquid nitrogen at −80°C until use. Patients did not receive adjuvant therapy or endocrine therapy before surgery. Clinical features of the samples are listed in . This study was approved by the ethics committee of The First Affiliated Hospital of Wenzhou Medical University. Written informed consent was obtained from all patients. Experiments involving human tissues were conducted in accordance with the Declaration of Helsinki.
Table 1 Clinicopathological features and hsa_circ_0072309 expression in 32 breast cancer patients
Cell culture and transfection
Human breast cancer cell lines MCF-7 and T47D were obtained from the cell bank of the Chinese Academy of Science, Shanghai and maintained at 37°C in 5% CO2/95% air in DMEM (Hyclone, Thermo Fisher Scientific, Waltham, MA, USA) containing 10% FBS, 100 units/mL penicillin, and 100 µg/mL streptomycin.
MCF-7 and T47D cells stably overexpressing hsa_ circ_0072309 were constructed as previously reported.Citation13 In brief, the coding sequence of hsa_circ_0072309 was inserted into the PLCDH-cir vector (Ribobio, Guangzhou, China). The lentiviral vector was constructed by Hanbio (Shanghai, China). Transfection was performed in accordance with the manufacturer’s instructions. MCF-7 and T47D cells were selected with 0.5 µg/mL puromycin (Sigma-Aldrich Co., St Louis, MO, USA) for 2 weeks. MiR-492 mimics (5’-AGGAC-CUGCGGGACAAGAUUCUU-3’) or negative controls were synthesized by Genepharma (Shanghai, China) and transfected into cell lines using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific) following the manufacturer’s instructions. Transfection efficiency was confirmed through qRT-PCR.
Cell viability assay
Cell viability was quantified with Cell Counting Kit-8 (CCK-8) (Beyotime, Jiangsu, China). MCF-7 and T47D cells were seeded in 96-well plates at the density of 2×103 cells per well. At different time points, 100 µL of serum-free culture medium and 10 µL of CCK-8 solutions were added to each well. The plates were then incubated at 37°C for 1 hour. Absorbance was measured on ELX-800 spectrometer reader (Bio-Tek Instruments, Winooski, VT, USA) at 450 nm.
Colony formation assay
A total of 1×103 cells were seeded in 6-well plates. The medium was changed every 3 days. The cells were cultured until most colonies comprised more than 50 cells. Then, the cells were fixed with paraformaldehyde for 30 minutes and stained with 0.1% crystal violet. Finally, the number of colonies was counted.
Flow cytometry analysis
Cells were plated in 12-well plates at the density of 1×106 cells per well. At 24 hours after incubation, the cells were fixed with ice-cold 70% ethanol and treated for 30 minutes with 1 unit of DNase-free RNase and 1 mg/mL propidium iodide at 37°C. Then, 2×104 cells were analyzed with Arial III FACS system (BD Biosciences, San Jose, CA, USA).
Luciferase assays
The possible target miRNAs of hsa_circ_0072309 were predicted using the Circinteractome tool (https://circinteractome.nia.nih.gov). Sequences containing potential wild-type or mutant binding sites for miRNAs in hsa_circ_0072309 were constructed into pmirGLO vectors (Promega Corporation, Fitchburg, WI, USA). For the luciferase reporter assay, the reporter vector and miRNAs were co-transfected into MCF-7 cells using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific). At 48 hours after induction, luciferase activity was measured using the dual-luciferase reporter assay system (Promega Corporation). Relative firefly luciferase activity was normalized to Renilla luciferase activity.
Migration and invasion assays
Migration and invasion assays were performed with chambers with pore sizes of 8 µm. For invasion assays, 3×104 cells were seeded into Matrigel-precoated chambers, and complete medium was added to the bottom of the wells to stimulate invasion. After 24 hours, the upper chambers were fixed with methanol and stained with crystal violet solution, and five fields (up, down, center, left, and right ×200) were selected for the quantification of invading cells. The experimental procedure of the migration assay was the same as that of the invasion assay. The migration assay, however, was not performed with precoated chambers.
qRT-PCR
RNA was isolated with TRIzol reagent (Thermo Fisher Scientific), precipitated with phenol–chloroform and ethanol, and then resuspended in RNase-free H2O. cDNA was prepared with PrimeScript RT reagent kit (TaKaRa, Japan) and subjected to qRT-PCR with ABI 7300 real-time PCR System (Applied Biosystems, Thermo Fisher Scientific). U6 and GAPDH were used as the internal controls.
Tumor xenograft assay
A total of 5×106 MCF-7 cells were subcutaneously injected into the right flanks of 4-week old female BALB/c (nu/nu) mice. A precision caliper was used to measure tumor size, which was recorded as the maximum diameter (a, mm) and vertical short diameter (b, mm). Tumor volume was calculated using the formula V (mm3) = 1/2abCitation2. At 4 weeks after injection, the mice were killed, and subcutaneous tumor weight was measured. Animal experiment protocols were approved by the Animal Ethics Committee of The First Affili ated Hospital of Wenzhou Medical University. All animal operations were performed in accordance with the Animal Policy and Welfare Committee of The First Affiliated Hospital of Wenzhou Medical University.
Statistical analyses
SPSS 19.0 software (IBM Corporation, Armonk, NY, USA) was used for data analysis, and all data from three independent experiments were shown as mean ± SD. Student’s t-test was used to evaluate differences, and P<0.05 was considered as a statistically significant difference.
Results
hsa_circ_0072309 was downregulated in breast cancer tissues
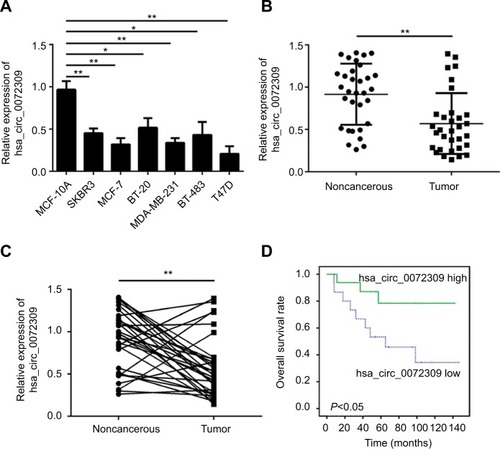
CircRNAs, a large class of mammalian RNAs, have regulatory potentialCitation10 and essential roles in cancer development.Citation9,Citation14 Nevertheless, the functions of circRNAs have not been fully studied. We first analyzed hsa_circ_0072309 expression in breast cancer cell lines to identify the function of hsa_circ_0072309 in breast cancer carcinogenesis. Our qRT-PCR results showed that hsa_ circ_0072309 expression was downregulated in all six breast cancer cell lines relative to that in the normal breast cell line (). We further analyzed hsa_circ_0072309 expression levels in breast cancer tumor tissues. Our analytical results for the 32 pairs of tumor tissues and adjacent normal tissues show that hsa_circ_0072309 expression was downregulated in tumor tissues (). Specifically, hsa_circ_0072309 expression was decreased in more than 70% of patients with tumors (). In line with the downregulation of hsa_ circ_0072309 in breast cancer samples, survival analysis also showed that hsa_circ_0072309 downregulation was correlated with poor survival rates (). All these results indicate that hsa_circ_0072309 was downregulated in breast cancer.
Figure 1 hsa_circ_0072309 was downregulated in breast cancer tissues.
Notes: (A) Relative expression of hsa_circ_0072309 in breast cancer cell lines was assessed by qRT-PCR. MCF-10A is a noncancerous control cell line. Other cell lines are cancerous. (B) Relative expression of hsa_circ_0072309 in 32 pairs of breast cancer tissues and adjacent noncancerous tissues. (C) Expression of hsa_circ_0072309 was downregulated in the 78.13% (25/32) breast cancer tissues compared to adjacent normal tissues. (D) Overall survival curve for 32 breast cancer patients with high or low hsa_circ_0072309 expression by Kaplan–Meier analysis. *P<0.05 and **P<0.01, vs control group. Every result was from three independent experiments.
hsa_circ_0072309 overexpression inhibited breast cancer progression
We then conducted functional assays to study the role of hsa_circ_0072309 in breast cancer carcinogenesis. We overexpressed hsa_circ_0072309 in MCF-7 and T47D breast cancer cell lines () and analyzed the proliferation rates of hsa_circ_0072309-overexpressing cancer cells. Our CCK-8 assay results show that hsa_circ_0072309 overexpres-sion inhibited MCF-7 and T47D cell proliferation (). This effect increased the number of cells in the G0/G1 stage (). Moreover, hsa_circ_0072309 overexpression reduced colony formation ().
Figure 2 Overexpression of hsa_circ_0072309 inhibited breast cancer progression.
Notes: (A) Cells were transfected with vector or oe-hsa_circ_0072309. Forty-eight hours after transfection, qRT-PCR analysis was performed to determine hsa_ circ_0072309 expression. (B) Cell Counting Kit-8 proliferation assay indicated that the proliferation in hsa_circ_0072309 overexpression group was stronger than that in control group. (C, D) Cell cycle analysis was conducted by flow cytometry. (E) Colony formation assay showed that hsa_circ_0072309 overexpression inhibited breast cancer cell proliferation. (F) Transwell migration assay showed that hsa_circ_0072309 overexpression inhibited breast cancer cell migration. (G) Transwell invasion assay indicated that hsa_circ_0072309 overexpression reduced the invaded cell numbers.*P<0.05, **P<0.01, and ***P<0.001 vs control group. Every result was from three independent experiments.
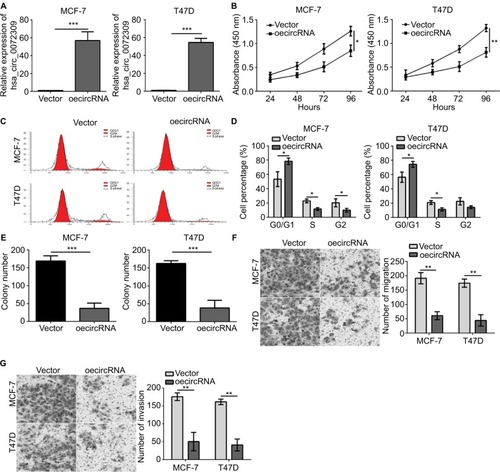
Given that migration and invasion capabilities are characteristics of malignant cancer cells, we determined whether hsa_circ_0072309 regulates the migration and invasion of breast cancer cells. The results of Transwell assays indicate that hsa_circ_0072309 overexpression inhibited the migration of MCF-7 and T47D cells (). In addition, the overexpression of hsa_circ_0072309 inhibited the invasion of MCF-7 and T47D cells ().
We knocked down hsa_circ_0072309 in MCF-7 and T47D cells to further investigate the physiological role of hsa_circ_0072309 (). The results of CCK-8 and Transwell assays show that hsa_circ_0072309 silencing enhanced the proliferation, migration, and invasion abilities of MCF-7 and T47D cells (). Thus, our data demonstrate that hsa_circ_0072309 overexpression suppressed breast cancer progression.
hsa_circ_0072309 regulated breast cancer progression by acting as the sponge of miR-492
CircRNAs function in cancer progression through several approaches, including sponging miRNA and regulating RNA transcription and protein production.Citation14 We first ana-lyzed the subcellular location of hsa_circ_0072309 to study the molecular mechanism underlying its function in breast cancer. We found that hsa_circ_0072309 was mainly located in the cytoplasm (). Thus, we hypothesized that hsa_circ_0072309 may function as an miRNA sponge. Then, we conducted bioinformatics analysis to predict the binding partners of hsa_circ_0072309 and identify the ten most probable target miRNAs of hsa_circ_0072309 (). We performed luciferase reporter assays to validate our predicted results. Our results illustrate that luciferase activity was most intensely downregulated by miR-492 overexpression in MCF-7 cells (). Notably, the results of our CCK-8 assay indicate that miR-492 overexpression resulted in the most drastic increase in the proliferation potential of MCF-7 cells (). Thus, we selected miR-492 for further investigation. Our qRT-PCR results show that hsa_circ_0072309 overexpression downregulated miR-492 expression (). Moreover, the deletion of the miR-492 binding site abrogated the suppressive effect of hsa_circ_0072309 on miR-492 expression in MCF-7 and T47D cells (). This effect indicates that hsa_circ_0072309 and miR-492 interaction would inhibit miR-492 expression. Additionally, miR-492 expression was upregulated in breast cancer tissues (). All of these results prove that hsa_circ_0072309 is a sponge of miR-492.
Figure 3 hsa_circ_0072309 was a sponge of miR-492 to regulate breast cancer progression.
Notes: (A) The levels of hsa_circ_0072309 were assessed by qRT-PCR in nuclear and cytoplasmic fractions. U6: nuclear control transcript; GAPDH: cytoplasmic control transcript. (B) Luciferase assays of MCF-7 cells co-transfected with indicated miRNAs and luciferase reporter containing hsa_circ_0072309-wild-type (WT) or mutant (Mut) construct. (C) CCK-8 assay was performed using MCF-7 cells. (D) Overexpression of hsa_circ_0072309 inhibited miR-492 expression in MCF-7 and T47D cells by qRT-PCR analysis. (E) MiR-492 expression was upregulated in breast cancer tissues compared to adjacent noncancerous tissues. (F) MiR-492 expression in MCF-7 and T47D cells transfected with oehsa_circ_0072309 and/or miR-492 mimic. (G) CCK-8 assay was performed to measure cell proliferation. (H) Cell cycle was determined by FACS analysis. (I, J) Transwell migration/invasion assays were utilized to test cell migration and invasion. *P<0.05 and **P<0.01 vs control group. Every result was from three independent experiments.
Abbreviation: CCK-8, Cell Counting Kit-8.
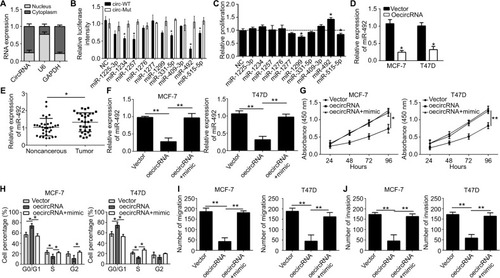
Then, we investigated whether hsa_circ_0072309 promotes breast cancer progression through miR-492. We rescued miR-492 expression in hsa_circ_0072309-overexpressing MCF-7 and T47D cells () and performed functional assays. Our results showed that the transfection of miR-492 mimics into hsa_circ_0072309-overexpressing cells rescued proliferation, migration, and invasion defects () and thus restored normal cell cycle progression (). These results indicate that hsa_circ_0072309 inhibited breast cancer progression through miR-492.
Table 2 Target miRNAs of hsa_circ_0072309
hsa_circ_0072309 overexpression inhibited breast cancer growth in vivo
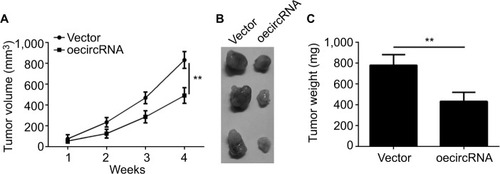
From the previously mentioned results, we learned that hsa_circ_0072309 inhibited breast cancer progression in vitro. Thus, we investigated whether hsa_circ_0072309 inhibited breast cancer progression in vivo. We injected hsa_circ_0072309-overexpressing cells into nude mice and measured tumor growth every week. Our results indicate that hsa_circ_0072309 overexpression decreased the growth () and sizes of breast cancer tumors (). These results show that hsa_circ_0072309 overexpression inhibited breast cancer progression in vivo.
Figure 4 hsa_circ_0072309 overexpression inhibited breast cancer growth in vivo.
Notes: (A) Tumor volume was monitored every week for 4 weeks. hsa_circ_0072309-overexpressing or control MCF-7 cells were injected into the flank of the nude mice recipients. (B, C) Tumor photo and weights were measured and presented at the endpoint of xenograft experiments. **P<0.01 vs control group. Every result was from three independent experiments.
Discussion
Information on the function of circRNAs in human disease remains limited. Similar to ncRNAs, circRNAs are expressed in multiple tissues and are highly diverse. Similar to tumor cells, different circRNAs exhibit distinctive expression patterns in various tissues and have specific functions in tumor progression. For example, circ_001895 is downregulated in gastric cancer tissue and cell lines and is closely correlated with gastric cell differentiation.Citation15 In contrast, circ_0000069 is upregulated in colorectal cancer and promotes colorectal cancer cell proliferation, migration, and invasion.Citation16 These findings indicate that circRNAs have important regulatory roles in tumorigenesis. In this study, we found that hsa_circ_0072309 expression was downregulated in breast cancer tissues and cells. We preliminarily determined the biological function of hsa_circ_0072309 through further analysis.
The expression of different circRNAs is associated with the pathogenesis of breast cancer.Citation17,Citation18 Circ-ABCB10 is upregulated in breast cancer and promotes breast cancer progression.Citation19 We discovered that hsa_circ_0072309 was downregulated in breast cancer tissues. Thus, the expression pattern of hsa_circ_0072309 opposes that of circ-ABCB10. In our study, we found that the downregulation of hsa_ circ_0072309 promoted the progression of breast cancer, and the expression of hsa_circ_0072309 was associated with the survival of breast cancer patients. Further functional assays showed that hsa_circ_0072309 overexpression inhibited MCF-7 and T47D cell proliferation, migration, and invasion. Thus, hsa_circ_0072309 functions as a tumor suppressor in breast cancer progression.
Similar to lncRNAs, circRNAs regulate miRNA expression in the cytoplasm by acting as miRNA sponges.Citation20,Citation21 The adsorption of circRNA to miRNA releases mRNAs from functional suppression by miRNA through reducing miRNA expression and would result in the formation of the cir-cRNA–miRNA–mRNA regulatory axis.Citation22 Cooperative communication between non-coding RNA and protein-coding RNA contributes to the biological complexities of cancer.Citation23 Circ-MTO1 can suppress the progression of hepatocellular carcinoma through the circ-MTO1–miR-9–p21 axis.Citation24 In the present study, we found that hsa_circ_0072309 inhibited breast cancer progression through miR-492 given that miR-492 overexpression in hsa_circ_0072309-overexpressing cells rescued the failure of breast cancer progression. A previous study suggested that mIR-492 promotes the proliferation, migration, and invasion of breast cancer cells.Citation25 Our study also demonstrated the oncogenic role of miR-492 in breast cancer.
Overall, we discovered that hsa_circ_0072309 is down-regulated in breast cancer. Moreover, hsa_circ_0072309 functions as a tumor suppressor in breast cancer by inhibiting cell proliferation, migration, and invasion via miR-492. We discovered a critical hsa_circ_0072309–miR-492 function axis in breast cancer progression. Nevertheless, the function of miR-492 in breast cancer progression requires further investigation.
Acknowledgments
We thank all patients involved in this study.
Supplementary material
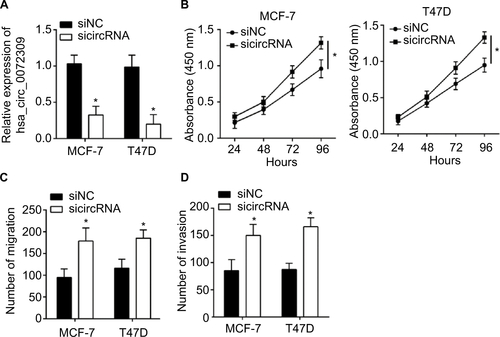
Figure S1 hsa_circ_0072309 silencing promotes breast cancer cell proliferation, migration, and invasion.
Notes: (A) qRT-PCR showed hsa_circ_0072309 level was decreased after siRNA transfection. (B) CCK-8 assay showed that hsa_circ_0072309 silencing promoted proliferation. (C, D) Transwell assay showed that hsa_circ_0072309 silencing led to enhanced migration and invasion in MCF-7 and T47D cells. *P<0.05.
Abbreviation: CCK-8, Cell Counting Kit-8.
Disclosure
The authors report no conflicts of interest in this work.
References
- ElfarGAEbrahimMAElsherbinyNMEissaLAValidity of Osteo-protegerin and Receptor Activator of NF-κB Ligand for the Detection of Bone Metastasis in Breast CancerOncol Res201725464165027983911
- ZhangZWangJGaoRDownregulation of MicroRNA-449 Promotes Migration and Invasion of Breast Cancer Cells by Targeting Tumor Protein D52 (TPD52Oncol Res201725575376127983918
- van DiestPJvan der WallEBaakJPPrognostic value of proliferation in invasive breast cancer: a reviewJ Clin Pathol200457767568115220356
- ZhangNCaoCZhuYPrimary breast lymphoma: A single center studyOncol Lett20171321014101828356993
- MengSZhouHFengZCircRNA: functions and properties of a novel potential biomarker for cancerMol Cancer20171619428535767
- NigroJMChoKRFearonERScrambled exonsCell19916436076131991322
- ChenJYHourTCYangSFAutophagy is deficient in nasal polyps: implications for the pathogenesis of the diseaseInt Forum Allergy Rhinol20155211912325533020
- ChenJLiYZhengQCircular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancerCancer Lett201738820821927986464
- KristensenLSHansenTBVenøMTKjemsJCircular RNAs in cancer: opportunities and challenges in the fieldOncogene201837555556528991235
- MemczakSJensMElefsiniotiACircular RNAs are a large class of animal RNAs with regulatory potencyNature2013495744133333823446348
- HansenTBJensenTIClausenBHNatural RNA circles function as efficient microRNA spongesNature2013495744138438823446346
- SuCHanYZhangHCiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-κB signallingJ Cell Mol Med20182263097310729532994
- YangCYuanWYangXCircular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expressionMol Cancer20181711929386015
- LiJYangJZhouPCircular RNAs in cancer: novel insights into origins, properties, functions and implicationsAm J Cancer Res20155247248025973291
- ShaoYChenLLuRDecreased expression of hsa_circ_001895 in human gastric cancer and its clinical significancesTumor Biol2017394101042831769912
- GuoJNLiJZhuCLComprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancerOnco Targets Ther201697451745828003761
- TangYYZhaoPZouTNCircular RNA hsa_circ_001982 Promotes Breast Cancer Cell Carcinogenesis Through Decreasing miR-143DNA Cell Biol2017361190190828933584
- LiuYLuCZhouYZhangZSunLCircular RNA hsa_ circ_0008039 promotes breast cancer cell proliferation and migration by regulating miR-432-5p/E2F3 axisBiochem Biophys Res Commun2018502335836329807010
- LiangHFZhangXZLiuBGJiaGTLiWLCircular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271Am J Cancer Res2017771566157628744405
- ChenKLvXLiWAutophagy Is a Protective Response to the Oxidative Damage to Endplate Chondrocytes in Intervertebral Disc: Implications for the Treatment of Degenerative Lumbar DiscOxid Med Cell Longev20172017985919
- DongYHeDPengZCircular RNAs in cancer: an emerging key playerJ Hematol Oncol2017101228049499
- RongDSunHLiZAn emerging function of circRNA-miRNAs-mRNA axis in human diseasesOncotarget2017842732717328129069868
- FaraziTAHorlingsHMTen HoeveJJMicroRNA sequence and expression analysis in breast tumors by deep sequencingCancer Res201171134443445321586611
- HanDLiJWangHCircular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progressionHepatology20176641151116428520103
- ShenFCaiWSFengZMiR-492 contributes to cell proliferation and cell cycle of human breast cancer cells by suppressing SOX7 expressionTumour Biol20153631913192125407488
