Abstract
The miR17-92 family is found to be aberrantly expressed and associated with clinicopathological characteristics in patients with various cancers, including digestive system cancers. However, its prognostic value is not yet established. Therefore, we performed a systematic review and meta-analysis to investigate the association between miR17-92-family expression and clinical outcomes in digestive system cancers. We searched the PubMed, Web of Science, Embase, and CNKI (Chinese) databases to retrieve eligible studies up to June 30, 2018. Prognostic data and clinicopathological features of overall survival (OS), disease-free survival (DFS), and progression-free survival (PFS) were extracted to evaluate correlations of the miR17-92 family with digestive system cancers. We used HRs to assess association between miR17-92-family expression and cancers prognosis. A total of 30 qualifying studies involving 4,056 subjects were included in this meta-analysis. Our results indicated that expression levels of miR17-92 can predict poor OS (HR 1.21, 95% CI 1.03–1.39; P=0). However, there was no relationship between the miR17-92 family and DFS (HR 0.86, 95% CI 0.6–1.11; P=0.170) or PFS (HR 1.37, 95% CI 0.83–1.91; P=0). Moreover, miR17-92 was related to TNM stage (III/IV vs I/II, HR 1.37, 95% CI 1.17–1.570; P=0.012), but there was no relationship between miR17-92 and metastasis (HR 1.64, 95% CI 1.34–1.95; P=0.491) or tumor size (≥5 cm vs <5 cm, HR 1.29, 95% CI 1.09–1.49; P=0.586). Subgroup analysis showed that miR17-92 expression was associated with poor OS among the Chinese subgroup (HR 1.28, 95% CI 1.08–1.48; P=0) and tissue samples (HR 1.12, 95% CI 0.93–1.31; P=0), while there was no association with other characteristics. Our results indicated that miR17-92 expression is significantly associated with poor survival in patients with digestive system cancers, suggesting that miR17-92 may be a promising prognostic marker to monitor prognosis and progression of cancers.
Introduction
Digestive system cancers include esophageal squamous-cell carcinoma (ESCC), gastric cancer (GC), gallbladder carcinoma, hepatocellular carcinoma (HCC), pancreatic cancer (PC), colorectal cancer (CRC), and colon cancer (CC). In spite of advanced development in clinical research in recent years, cancer is still the main factor in death worldwide. It is estimated that ~1.7 million people were diagnosed with cancers and 0.6 million people died of malignancies in the US in 2017.Citation1 High morbidity and mortality rates in digestive system cancers remain an important health problem in developing countries.Citation2 Due to the lack of understanding of the molecular mechanisms of cancer, it is difficult to identify a reliable biomarker to detect cancer and find an effective therapeutic agent for clinical application. Several independent factors can be used to identify and evaluate the clinical outcomes of human cancers, consisting of depth of tumor invasion, histological grade, TNM stage, and metastasis to lymph nodes.Citation3 Pathological biopsy is still the gold standard to diagnose diseases, but is an invasive method and needs high requirements for technology. Blood-based tumor bio-markers are used widely to diagnose cancers and predict the prognosis of neoplasms. However, because of low sensitivity and specificity, this detection method is far from satisfactory.Citation4 Therefore, it is essential to find a less invasive and more accurate marker to apply to clinical medicine urgently.
In recent decades, a number of studies have found that miRNA, which belongs to a class of RNA transcripts 20–22 nucleotides in length without a protein-coding function, is closely related to tumor development and progression.Citation5 According to mRNA degradation and translational repression, miRNA can regulate gene expression posttranscriptionally by binding to the 3’-untranslated region of target mRNAs.Citation6 The earliest proof of miRNA involvement in human cancer was provided by Calin et al from studies attempting to identify tumor suppressors at chromosome 13q14 in B-cell chronic lymphocytic leukemia cells.Citation7 In recent decades, numerous articles have indicated that aberrant expression of miRNA in human cancers is related to many processes of tumorigenesis, including cell proliferation, differentiation, angiogenesis, and metastasis.Citation5–Citation7 Cancer cells with abnormal miRNA-expression levels evolve the property to sustain proliferative signaling, evade growth suppressors, resist cell death, activate invasion and metastasis, and induce angio-genesis.Citation8 Roles of miRNAs in human cancers are examined from the viewpoint of dysregulation. Oncogenic miRNAs are involved in the overexpression of cancers, whereas suppressive miRNAs are involved in the downregulation of cancers.Citation9 Because of these fundamental activities, miRNAs have been proven to act as tumor oncogenes or suppressors.Citation10 In addition, miRNA is stable in circulation (such as whole blood, plasma, serum, sputum) and formalin-fixed paraffin-embedded tissue; therefore, it is regarded as a biomarker for cancer diagnosis and prognosis.Citation11
The miR17-92 family, located at human chromosome 13q31, is one of the most extensively studied miRNA clusters and has been shown to play important roles in the pathogenesis of various cancers, including glioma,Citation12 Burkitt’s lymphoma,Citation13 lung cancer,Citation14 osteosarcoma,Citation15 and digestive system cancers. The miR17-92 family has six members (miR17, miR18a, miR19a, miR20a, miR19b, and miR92a) and two paralogues (miR106a and miR106b).Citation16 High expression of the miR17-92 cluster promotes the metastasis of cancers, indicating its role as an oncogene.Citation15 However, studies have suggested that miR17-5p can inhibit metastasis and invasion of tumors.Citation17 Wang et alCitation18 found that higher expression levels of miR17-5p/20a were significantly correlated with poor overall survival (OS). Xue et alCitation19 found that the OS of patients was negatively associated with high levels of miR20b in GCs, but other studies have demonstrated the contrary role of the miR17-92 cluster in cancer outcomes. Fan et alCitation20 identified that patients with lower expression levels of miR20a had significantly poor recurrence-free survival and OS in HCC patients. Therefore, the role of miR17-92 in cancer development and the exact mechanism are not yet consistent.
Previous discrepant results may due to several factors, including sample size, race, detection method, and tumor metastasis. As such, further studies are needed to evaluate the association between the expression of miR17-92 with the prognosis of cancers. A lot of articles have indicated that a similar sequence of miRNAs may regulate a group of target mRNAs and a set of biomarkers may be a better indicator than a single one. Therefore, we conducted this systematic review and meta-analysis to explore the clinical significance of the miR17-92 family as prognostic markers in human digestive system cancers.
Methods
Search strategy
A comprehensive search was performed on the PubMed, Web of Science, Embase, and CNKI (Chinese) databases for articles published to June 30, 2018. Search terms used were “miR17-3p” OR “miR17-5p” OR “miR18a” OR “miR19a” OR “miR19b” OR “miR20a” OR “miR92a” OR “miR106a” OR “miR106b”, “carcinoma” OR “cancer” OR “tumor” OR “malignancy” OR “neoplasia” OR “sarcoma”, “prognosis” OR “prognostic”, “outcome”, and “survival”. In addition, we attempted to find other potential available studies by searching the references and relevant published articles manually. Because this is a systematic review and meta-analysis, ethical approval and patient written informed consent were not required.
Inclusion and exclusion criteria
All eligible studies were reviewed and evaluated based on PRISMA.Citation21 Inclusion criteria were: expression of miR17-92 detected in digestive system cancers; study based on human research; cohort or case–control study; sufficient data to retrieve HRs for survival and corresponding 95% CIs; and published in Chinese or English. We excluded studies if they were conducted on animals or cell lines, were reviews, letters, case reports, conference meetings, or comments, were duplicate publications, or did not provide or had no available data to calculate HRs and 95% CIs. The quality of the included studies was evaluated and examined by the authors after browsing the abstracts and full texts of manuscripts. The final decision was reached by discussion.
Data extraction
According to the inclusion and exclusion criteria, data were extracted by two investigators (PX and RZ) independently. If an article potentially qualified for the meta-analysis, the full text of the study was required. Any discrepancies were resolved by discussion and consensus. Information extracted was first author’s name, publication year, country, total number of patients, cancer type, specimen source, detection method, follow-up time, cutoff value, TNM stage, metastasis, tumor size, HRs with 95% CIs for OS and cancer progression, including disease-free survival (DFS), progression-free survival (PFS), recurrence-free survival, disease-specific survival, cancer-specific survival, cancer-free survival, and event-free survival. All HRs and 95% CIs were extracted from the original literature. If survival data were provided i only n a Kaplan–Meier curve, HRs with 95% CIs were digitized and extracted using Engauge Digitizer (version 4.1) software, designed by Tierney et al.Citation22 Data were extracted from multivariate analyses if both univariate and multivariate results were provided in the same study.
Quality assessment
The quality of the included studies was systematically evaluated by two investigators based on the Newcastle–Ottawa Scale standard,Citation23 which included three parts: selection (4 points), comparability (2 points), and outcome (3 points). Scores range from 0 to 9 points. A study with a score ≥6 points was regarded as high quality.
Statistical analysis
All extracted data were analyzed using Stata software version 12.0 (StataCorp, College Station, TX, USA). Pooled HRs with 95% CIs for OS, DFS and PFS were calculated to estimate the association between miR17-92 expression and prognosis of digestive system cancers. Cochran’ s Q test and Higgins’s I2 statistic were used to evaluate heterogeneity among the selected studies. If P<0.1 or I2>50%, it indicated that heterogeneity existed, and a random effect-model would be used; otherwise, a fixed-effect model would be applied (P>0.1 or I2<50%). Subgroup analysis was conducted to investigate potential heterogeneity based on country, cancer type, and tumor size. Sensitivity analysis was performed to explore the influence of single studies by omitting one study at a time. Additionally, publication bias was assessed using Begg’s and Egger’s tests. P<0.05 was considered statistically significant.
Results
Data selection and study characteristics
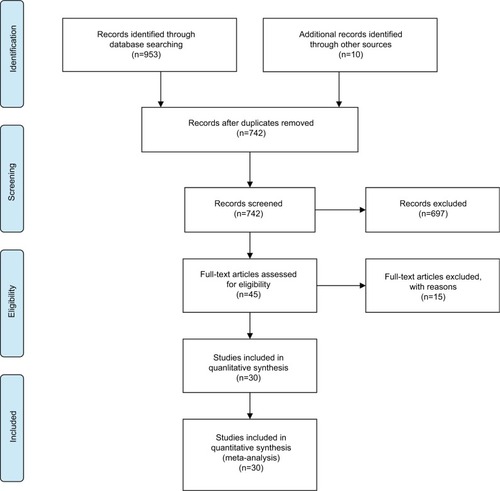
A total of 963 articles were retrieved from the online databases and other sources in accordance with the search strategies. After removal of duplicates, there existed 742 studies. After screening of titles and abstracts, 697 were considered ineligible. A total of 45 potential articles were carefully reviewed via full text, and then 15 studies were excluded due to a lack of sufficient data. Finally, 30 eligible studiesCitation18–Citation20,Citation24–Citation50 were included in our meta-analysis. The selection flowchart for this meta-analysis is shown in .
The main characteristics and quality of the eligible studies (2008–2018) assessed with the Newcastle–Ottawa Scale are summarized in . Among these 30 articles, 21 were from China, four from Japan, two from Spain, one from the US, one from South Korea, and one from Turkey. A total of 4,056 patients were included in our meta-analysis, with a maximum sample size of 735 and a minimum sample size of 22 participants. The type of digestive system cancers included ESCC, GC, HCC, hepatoblastoma, PC, CRC, and CC. HRs with 95% CIs were extracted directly from 26 original studies,18–20,24,26–28,30–38,40–49 while two articlesCitation25,Citation50 provided only survival curves, so we indirectly calculated the values from Kaplan–Meier curves based on the method proposed by Tierney.Citation22 Two studiesCitation29,Citation39 provided the risk ratios only; therefore, we merged HRs and risk ratios. Quantitative real-time PCR was used to detect miR17-92 expression in 29 studies, while one article used microarrays. Sample types included tissue, serum, and plasma. Because of the variations in cutoff definitions, cutoff values were different in these studies.
Table 1 Main characteristics of all included studies
Association between miR17-92 expression and clinicopathological features
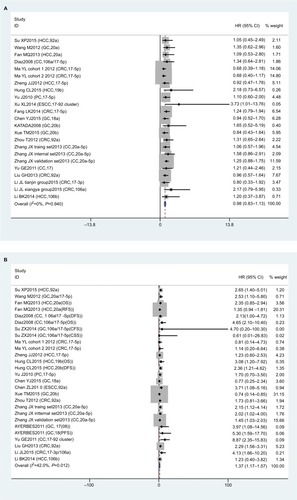
Meta-analysis indicated that there were 23 studies reporting on correlations of miR17-92-expression with sex; however results showed that expression levels were not associated with sex (HR 0.98, 95% CI 0.83–1.13; P=0.940; ). There were 27 studies on TNM stage (III/IV vs I/II), and values suggested that miR17-92 expression was linked with advanced TNM stage (HR 1.37, 95% CI 1.17–1.57; P=0.012; ). In addition, 24 articles investigated the association between miR17-92 expression and distant metastasis and 12 studies tumor size (≥5 cm vs <5 cm). However, no significance was found for distant metastasis (P=0.491, ) or tumor size (P=0.586, ). Due to the lack of sufficient data, we did not analyze any association between miR17-92 family expression and other clinicopathological characteristics.
miR17-92 expression and OS
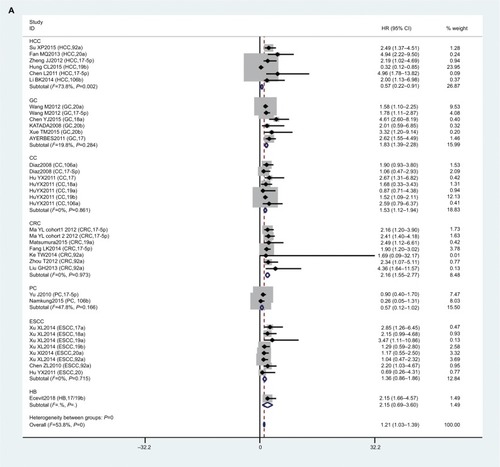
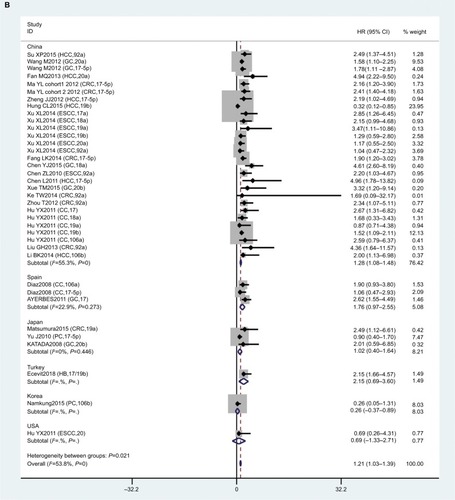
In this meta-analysis, 23 studies reported that expression levels of the miR17-92 family were related to OS. HRs with 95% CIs were retrieved from these articles. We found that miR17-92-expression levels were related to poor OS in digestive system patients (HR 1.21, 95% CI 1.03–1.39; P=0; ).
Figure 3 Forest plot of association between miR17-92 family and overall survival for digestive system cancers.
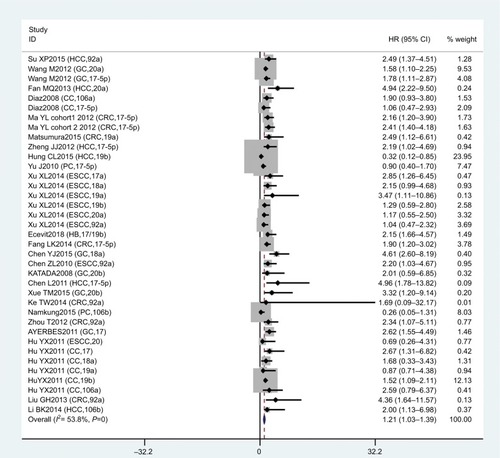
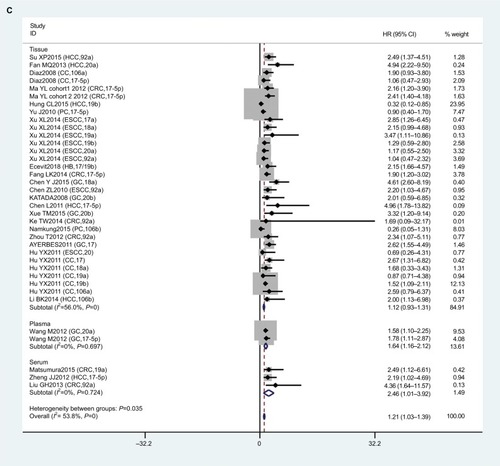
To reduce the effect of heterogeneity, we performed a subgroup analysis based on country (), cancer type (), and sample source (). We observed that miR17-92 had a great influence on OS in the HCC subgroup (HR 0.57, 95% CI 0.22–0.91; P=0.020), but no significant effect on GC (P=0.284), CC (P=0.861), CRC (P=0.973), ESCC (P=0.715), or PC (P=0.166). Then, we detected a significant association between miR17-92 expression and poor OS in patients with cancer in the China subgroup (HR 1.28, 95% CI 1.08–1.48; P=0); however, there was no significant effect in the Spain (P=0.273) or Japan (P=0.446) subgroups. Finally, we conducted a subgroup analysis on sample source and found that miR17-92 was strongly related to tissue samples (HR 1.12, 95% CI 0.93–1.31; P=0), while there was no influence on plasma (P=0.697) or serum (P=0.724) samples.
miR17-92 expression and DFS
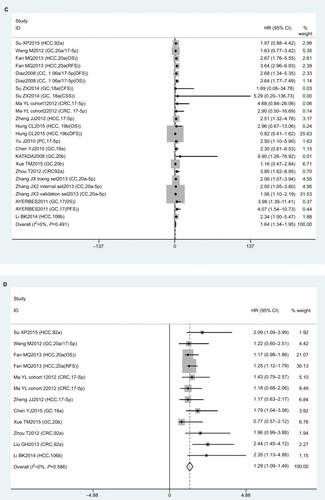
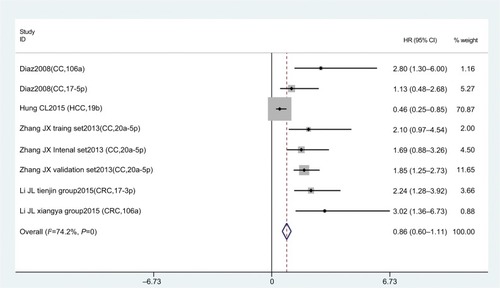
Eight studies focused on DFS analysis. Due to the relatively high heterogeneity value (I2=74.2%), we used a random-effect model to calculate HRs and 95%CIs for DFS. The results showed that there was no statistical association between miR17-92 expression and cancer DFS (HR 0.86, 95% CI 0.60–1.11; P=0; ).
miR17-92 expression and PFS
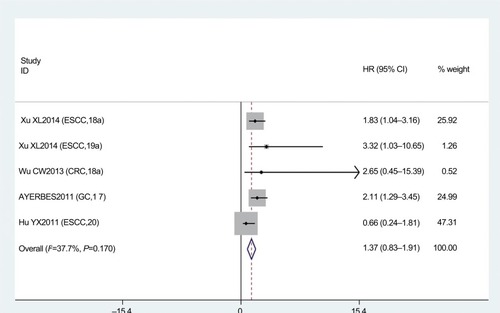
PFS analysis was done in five studies. No association was found between miR17-92 expression and cancer PFS (HR 1.37, 95% CI 0.83–1.91; P=0.170; ). Because only five articles evaluated PFS, this was too small a sample to conduct subgroup analysis for PFS.
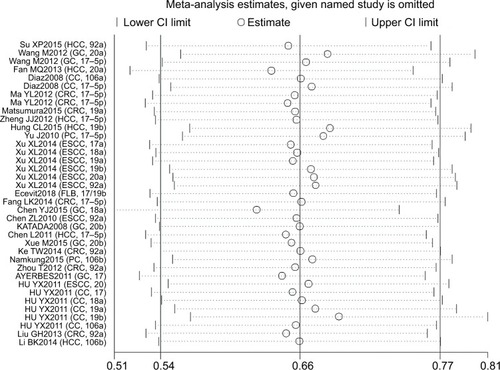
Sensitivity analysis and publication bias
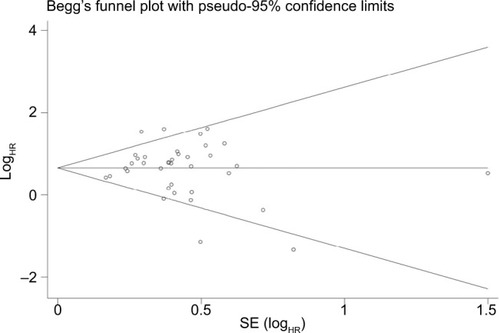
Sensitivity analysis was conducted with Stata 12.0 to evaluate whether any individual study influenced the overall consequences. The results showed that any single article had little effect on final values (). We used Begg’s and Egger’s funnel plots to evaluate the publication bias of the studies included in our meta-analysis, and results indicated that there was no significant publication bias in the pooled analysis of cancer prognosis ().
Discussion
The occurrence and development of cancers are multifactorial, multistep, and complicated processes.Citation1 Due to the lack of early diagnostic and prognostic indices, lots of patients are diagnosed in the advanced stage. Recently, many studies have demonstrated that miRNAs play important roles in the tumorigenesis of various cancers, including angiogenesis, cell proliferation, differentiation, invasion, apoptosis, and metastasis.Citation5,Citation6 As such, miRNAs have been considered oncogenes and cancer suppressors. It is hypothesized that miRNAs can regulate more than a third of eukaryotic genes.Citation9 Exploring the functions of miRNAs and their target genes involved in biogenesis may improve understanding of the potential mechanisms of tumor procession and offer significant insights into the diagnosis, prognosis, and treatment of cancers.Citation12,Citation13 Published results have indicated that miRNAs are stable in circulation and tissue and can be promising noninvasive biomarkers for diagnosis and prognosis of cancers.Citation17
Previous research has concluded that aberrant expression of miR17-92 is relevant in cancer development, includ ing Burkitt’s lymphoma,Citation13 breast cancer,Citation17 and GC.Citation18 Many studies have demonstrated that high miR17-92 expression indicates poor clinical characteristics and worse prognosis in digestive system cancers; however, results have not reached agreement as yet. Therefore, we performed the first meta-analysis of eligible studies to evaluate systematically the prognostic significance of miR17-92 in digestive system cancers. In our meta-analysis, the results suggested that high expression levels of miR17-92 represented a risk factor for poor OS (HR 1.21, 95% CI 1.03–1.30; P=0) in digestive system cancers. This demonstrates that the miR17-92 family could be indicators for the prognosis of cancers. Unfortunately, there was no association between miR17-92 expression and DFS (HR 0.86, 95% CI 0.60–1.11; P=0.000) or PFS (HR 1.37, 95% CI 0.83–1.91; P=0.170) in this meta-analysis. Moreover, investigating the effect of pathological features on OS, we found that high expression levels of miR17-92 were significantly associated with TNM stage (III/IV vs I/II, HR 1.37, 95% CI 1.17–1.57; P=0.012), but there was no correlation with metastasis (P=0.491) or tumor size (P=0.586).
In addition, we conducted subgroup analyses to explore the prognostic value of miR17-92 in OS and have successfully acquired some valuable conclusions for clinical application. Results showed that miR17-92 levels predicted poor prognosis in the China subgroup (HR 1.28, 95% CI 1.08–1.48; P=0.000), but not in Spain (P=0.273) or Japan (P=0.446). This diversity may be due to geographical locations, ethnicity, climate, and different lifestyles. Meanwhile, to evaluate the relationship between miR17-92 expression and prognosis based on sample sources, we performed subgroup analyses and found that the tissue subgroup had poor OS (HR 1.12, 95% CI 0.93–1.31; P=0), but no such relationship was found with serum (P=0.724) or plasma (P=0.697). Furthermore, we also noticed that miR17-92 expression was associated with a favorable prognosis in HCC (HR 0.57, 95% CI 0.22–0.91; P=0.02). Then, sensitivity analyses were carried out to assess whether the heterogeneity of data had an effect on results. After removal one study at a time, there was no significant influence on the final outcome. This demonstrated that our results were relatively stable and credible. Also, publication bias did not reach statistical significance.
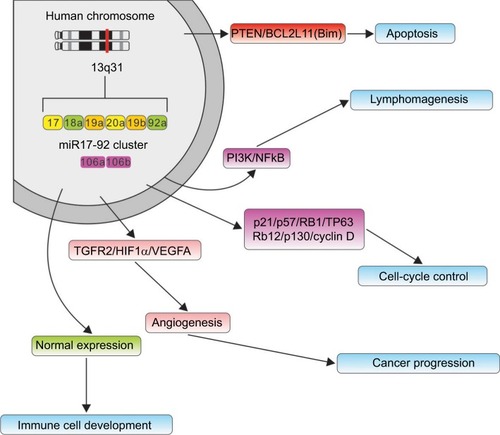
The miR17-92 cluster was the first miRNA gene implicated in human cancers. However, the potential mechanisms of the miR17-92 cluster in cancer prognosis have not been fully elucidated. Some researchers have demonstrated that miR17-92 functions may be connected with changes in cancer-related proteins and pathways ().Citation51,Citation54 Jung et alCitation52 globally investigated Ago2-bound mRNAs and found that miR17-92 obviously repressed numerous targets involved in the instability of mRNA, while the miRNAs repressed expression of their targets, enhanced stability, and lengthened the poly-A tails of nontarget mRNAs. Additionally, the expression of miR17-92 was negatively associated with expression of BTG3, TOB1, CSNK1A1, and ANKRD52 in cancer cell lines. Yang et alCitation53 demonstrated that up-regulation of miR17-92 contributed to the downregulation of QKI2 expression, and then, by decreasing the expression of β-catenin, they inhibited the proliferation, migration and invasion of tumor cells. All these results suggest that miR17-92 can promote tumorigenesis not only by posttranscriptionally increasing global gene expression but also by repressing downstream molecules.
Strength and limitations
It should be stressed that there are limitations in our meta-analysis. First, most of the included studies were from Asia, while European articles were few in number, which may have been an important source of the heterogeneity and inconsistent results found in our meta-analyses. This emphasizes the need for future studies to test the association of miR17-92 with prognosis in digestive system cancers in Western countries. Second, not all digestive system cancers were included in this meta-analysis. Third, some dataCitation25,Citation50 using HRs with 95% CIs were extracted from Kaplan–Meier survival curves, which inevitably brought tiny errors. Fourth, there was no uniform cutoff value to estimate expression levels of miR17-92, and actual values may have been in disagreement due to different algorithms and resulted in some heterogeneity. Finally, there were insufficient data to completely investigate the association between miR17-92 and clinicopathological characteristics of cancers, which needs more studies. Although with some limitations, our study is a comprehensive update, review, and meta-analysis focusing on the correlation of aberrant miR17-92 expression with the development and prognosis of digestive system cancers, providing new insight into the pathogenesis of digestive system cancers.
Conclusion
This systematic review and meta-analysis primarily investigated the expression of miR17-92 and clinical outcomes of patients with digestive system cancers. miR17-92 expression was associated with TNM stage in cancers. Our results demonstrated that the miR17-92 family might be promising prognostic biomarkers for digestive system cancers. Considering the limitations of this meta-analysis, further large-scale and high-quality prospective studies should be performed to validate these findings before clinical guidance using miR17-92 in the prognosis of cancers.
Author contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- SiegelRLMillerKDJemalACancer statistics, 2017CA Cancer J Clin201767173028055103
- FerlayJSoerjomataramIDikshitRCancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012Int J Cancer20151365E359E38625220842
- SvoronosAAEngelmanDMSlackFJOncomiR or tumor suppressor? The Duplicity of microRNAs in cancerCancer Res201676133666367027325641
- Perez-GraciaJLSanmamedMFBoschAStrategies to design clinical studies to identify predictive biomarkers in cancer researchCancer Treat Rev201753799728088073
- LeichterALSullivanMJEcclesMRChatterjeeAMicroRNA expression patterns and signalling pathways in the development and progression of childhood solid tumoursMol Cancer20171611528103887
- ChangCCYangYJLiYJCorrigendum to “MicroRNA-17/20a functions to inhibit cell migration and can be used a prognostic marker in oral squamous cell carcinoma” [Oral Oncol. 49(9) (2013) 923-931]Oral Oncol20177220220328711305
- CalinGADumitruCDShimizuMFrequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lym-phocytic leukemiaProc Natl Acad Sci USA20029924155241552912434020
- PengYCroceCMThe role of microRNAs in human cancerSignal Transduct Target Ther2016111500429263891
- Valencia-SanchezMALiuJHannonGJParkerRControl of translation and mRNA degradation by miRNAs and siRNAsGenes Dev200620551552416510870
- TakasakiSRoles of microRNAs in cancers and developmentMethods Mol Biol2015121837541325319665
- BobbiliMRMaderRMGrillariJDellagoHOncomiR-17-5p: alarm signal in cancer?Oncotarget2017841712067122229050357
- LuSWangSGengSMaSLiangZJiaoBIncreased expression of microRNA-17 predicts poor prognosis in human gliomaJ Biomed Biotechnol2012201261621836813
- RobainaMCFaccionRSMazzoccoliLMiR-17-92 cluster components analysis in Burkitt lymphoma: overexpression of miR-17 is associated with poor prognosisAnn Hematol201695688189127044389
- ChenQSiQXiaoSPrognostic significance of serum miR-17-5p in lung cancerMed Oncol201330135323263848
- LiXYangHTianQLiuYWengYUpregulation of microRNA-17-92 cluster associates with tumor progression and prognosis in osteosarcomaNeoplasma2014610445346024645838
- FedeliMRibaMGarcia ManteigaJMmiR-17~92 family clusters control iNKT cell ontogenesis via modulation of TGF-β signalingProc Natl Acad Sci USA201611351E8286E829527930306
- LiJLaiYMaJmiR-17-5p suppresses cell proliferation and invasion by targeting ETV1 in triple-negative breast cancerBMC Cancer201717174529126392
- WangMGuHWangSCirculating miR-17-5p and miR-20a: molecular markers for gastric cancerMol Med Rep201256151422406928
- XueTMTaoLDZhangMXuGCZhangJZhangPJmiR-20b overexpression is predictive of poor prognosis in gastric cancerOnco Targets Ther201581871187626244024
- FanMQHuangCBGuYXiaoYShengJXZhongLDecrease expression of microRNA-20a promotes cancer cell proliferation and predicts poor survival of hepatocellular carcinomaJ Exp Clin Cancer Res20133212123594563
- KnoblochKYoonUVogtPMPreferred reporting items for systematic reviews and meta-analyses (PRISMA) statement and publication biasJ Craniomaxillofac Surg2011392919221145753
- TierneyJFStewartLAGhersiDBurdettSSydesMRPractical methods for incorporating summary time-to-event data into meta-analysisTrials2007811617555582
- WellsGThe Newcastle-Ottawa scale (NOS) for assessing the quality of nonrandomised studies in meta-analysesAppl Eng Agric2014186727734
- SuXWangHGeWAn in vivo method to identify microRNA targets not predicted by computation algorithms: p21 targeting by miR-92a in cancerCancer Res201575142875288526062558
- ÇEAktaşSTosun YildirimHMicroRNA-17, microRNA-19b, microRNA-146a, MicroRNA-302d expressions in hepatoblastoma and clinical importanceJ Pediatr Hematol Oncol2018411712
- DíazRSilvaJGarcíaJMDeregulated expression of miR-106a predicts survival in human colon cancer patientsGenes Chromosom Cancer2010479794802
- SuZXZhaoJRongZHWuYGGengWMQinCKDiagnostic and prognostic value of circulating miR-18a in the plasma of patients with gastric cancerTumour Biol20143512121191212525416437
- MaYZhangPWangFElevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene p130Nat Commun201231129123250421
- MatsumuraTSugimachiKIinumaHExosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancerBr J Cancer2015113227528126057451
- ZhengJDongPGaoSWangNYuFHigh expression of serum miR-17-5p associated with poor prognosis in patients with hepatocellular carcinomaHepatogastroenterology20136012354955223108086
- HungCLYenCSTsaiHWSuYCYenCJUpregulation of microRNA-19b predicts good prognosis in patients with hepatocellular carcinoma presenting with vascular invasion or multifocal diseaseBMC Cancer201515166526453548
- YuJOhuchidaKMizumotoKFujitaHNakataKTanakaMMicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasionCancer Biol Ther201010874875720703102
- XuXLJiangYHFengJGSuDChenPCMaoWMMicroRNA-17, microRNA-18a, and microRNA-19a are prognostic indicators in esophageal squamous cell carcinomaAnn Thorac Surg20149731037104524360091
- FangLLiHWangLMicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expressionOncotarget20145102974298724912422
- WuCWDongYJLiangQYMicroRNA-18a attenuates DNA damage repair through suppressing the expression of ataxia telangiec-tasia mutated in colorectal cancerPLoS One201382e5703623437304
- ChenYJWuHZhuJMMicroRNA-18a modulates p53 expression by targeting IRF2 in gastric cancer patientsJ Gastroenterol Hepatol201631115516326173586
- ChenZLZhaoXHWangJWmicroRNA-92a promotes lymph node metastasis of human esophageal squamous cell carcinoma via E-cadherinJ Biol Chem201128612107251073421148309
- KatadaTIshiguroHKuwabaraYmicroRNA expression profile in undifferentiated gastric cancerInt J Oncol200934253754219148490
- ChenLJiangMYuanWTangHmiR-17-5p as a novel prognostic marker for hepatocellular carcinomaJ Invest Surg201225315616122583011
- KeTWWeiPLYehKTChenWTChengYWMiR-92a promotes cell metastasis of colorectal cancer through PTEN-mediated PI3K/Akt pathwayAnn Surg Oncol20152282649265525515201
- NamkungJKwonWChoiYMolecular subtypes of pancreatic cancer based on miRNA expression profiles have independent prognostic valueJ Gastroenterol Hepatol20163161160116726644397
- ZhouTZhangGLiuZXiaSTianHOverexpression of miR-92a correlates with tumor metastasis and poor prognosis in patients with colorectal cancerInt J Colorectal Dis2013281192422772712
- ZhangJXSongWChenZHPrognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysisLancet Oncol201314131295130624239208
- Valladares-AyerbesMBlancoMHazMPrognostic impact of disseminated tumor cells and microRNA-17-92 cluster deregulation in gastrointestinal cancerInt J Oncol20113951253126421743960
- HuYCorreaAMHoqueAPrognostic significance of differentially expressed miRNAs in esophageal cancerInt J Cancer2011128113214320309880
- YuGTangJQTianMLPrognostic values of the miR-17-92 cluster and its paralogs in colon cancerJ Surg Oncol2012106323223722065543
- LiuGHZhouZGChenRSerum miR-21 and miR-92a as bio-markers in the diagnosis and prognosis of colorectal cancerTumour Biol20133442175218123625654
- LiJLiuYWangCSerum miRNA expression profile as a prognostic biomarker of stage II/III colorectal adenocarcinomaSci Rep2015511292126250939
- LiBKHuangPZQiuJLLiaoYDHongJYuanYFUpregulation of microRNA-106b is associated with poor prognosis in hepatocellular carcinomaDiagn Pathol2014922625466449
- KomatsuSIchikawaDTsujiuraMPrognostic impact of circulating miR-21 in the plasma of patients with gastric carcinomaAnticancer Res201333127123267156
- JinHYLaiMXiaoCmicroRNA-17~92 is a powerful cancer driver and a therapeutic targetCell Cycle201413449549624419145
- JungESeongYJeonBKwonYSSongHMicroRNAs of miR-17-92 cluster increase gene expression by targeting mRNA-destabilization pathwaysBiochim Biophys Acta Gene Regul Mech20181861760361229935344
- YangHPengZLiangMThe miR-17-92 cluster/QKI2/β-catenin axis promotes osteosarcoma progressionOncotarget2018938252852529329861871
- GeorgeKChao-YiWHuang-YuYMiR-17-92 cluster and immunityJ Formos Med Assoc2018pii: S0929-6646183001630020