Dear editor
An article titled, “Prognostic values of HE4 expression in patients with cancer: a meta-analysis”, was recently published in the journal, Cancer Management and Research, detailing a study conducted by Dai et al.Citation1 This study aims to validate and estimate the effectiveness of HE4 as a prognostic marker in cancer. We commend the authors for the study’s focus on all types of cancers, as well as the associated subgroup analysis detailing the results for individual cancer types and population groups. This study is valuable as a guidepost for informing future research on HE4 as a prognostic biomarker to predict the cancer patients’ clinical outcomes. However, there are a few points that require to be addressed regarding the study conducted.
Promising biomarker in epithelial cancer diagnosis and prognosis
It is worth mentioning that HE4 is not only a possible prognostic marker, with recent studies showing that it is also a promising serum and tissue biomarker for diagnosis in lung cancer, ovarian carcinoma, and endometrial cancer, as well as a differential marker for identification of ovarian cancer from benign gynecologic disease.Citation2,Citation3 It is emerging as a clinically significant biomarker in ovarian cancer.
Systematic review and meta-analysis guidelines and protocol registration
Furthermore, the authors have not specified the following of any systematic review and meta-analysis guidelines. These guidelines (such as PRISMA, JBI, and MOOSE) exist to homogenize and standardize systematic reviews and meta-analysis to inform and benefit future research, which is one of the primary impetus behind conducting of a study, which cumulates and analyzes previous existing literature. It is also suggested that the protocols used for performing a systematic review and meta-analysis are registered in the PROSPERO database, which allows peers to replicate the type of study undertaken as well as provide insight on the process behind the study.
Publication bias of the included studies
It is also worth noting that the evaluation publication bias consists of a few key modules to evaluate the bias of the included studies. Although the Begg’s and Eggers’ test alongside the funnel plots were used for assessing publication bias in Dai et al’s study, another key assessment that is worth including is the “Duval and Tweedie’s trim and fill” method. This method is an essential module that may be used to impute missing small studies with large effect size to be dispersed equally on either side of the overall effect.Citation4
Survival endpoints
As this study considers disease-free survival (DFS) as an endpoint for its meta-analysis, an evaluation of recurrence rates is also possible as patients that do not fall under DFS would have suffered relapses. This additional analysis is a natural extension of the data already being presented, and might serve to help future research in this field.
The pooled effect size of cancer patients’ survival
Additionally, the pooled HRs for the subgroups of Asian (2.62) and lung cancer (2.31) as well as for DFS of all studies (2.50) are relatively high when compared to other subgroup cohorts and survival endpoints, as they indicate that the probability of death is over twice as likely for the patients expressing HE4 as compared to patients not expressing HE4 in cancer. These marked differences in HR values between different cancers and different populations require further evaluation and analysis.
Threshold effects: correlation between the ranks of effect sizes and the ranks of their variances
It is also notable that χ2 and I2 values may not be sufficient as statistical parameters as they do not consider the threshold effect. In random-effects meta-analysis, it may be useful to also include the τ2 parameter for estimating variance or heterogeneity between the effects for test accuracy, as the τ2 parameter does consider the threshold effect.Citation5
These points mentioned above are to address improvements that could benefit Dai et al’s study and help elevate its impact on future research. These points should also help guide future similar systematic reviews and meta-analyses.
Disclosure
The authors report no conflicts of interest in this communication.
Dear editor
We thank Dr Rama Jayaraj and Chellan Kumarasamy for their interest and comments. They came up with several valuable points that improved our meta-analysis.Citation1
As they mentioned, recent studiesCitation2,Citation3 showed that HE4 was a promising serum and tissue biomarker for diagnosis in lung cancer, ovarian carcinoma, and endometrial cancer, but its prognostic value remains controversial. Therefore, this is why we did this study to accurately assess the prognostic value of HE4 expression in cancer patients.
Our meta-analysis is based on Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines.Citation4 However, the review protocol was not registered in the PROSPERO database, which is an oversight of our work.
In terms of publication bias of the included studies, we think that the use of Begg’s and Eggers’ test alongside the funnel plots was enough to assess the publication bias. What’s more important, the “Duval and Tweedie’s trim and fill” methodCitation5 was currently controversial in assessing publication bias. So we did not use this method at that time. Now we can provide the result here. The combined HR and 95% CI of filled meta-analysis is 1.645 and 1.352–2.002 (random-effects model) ().
Figure 1 Filled funnel plots of publication bias for all of the included studies reported with overall survival.
Abbreviation: SE, standard error.
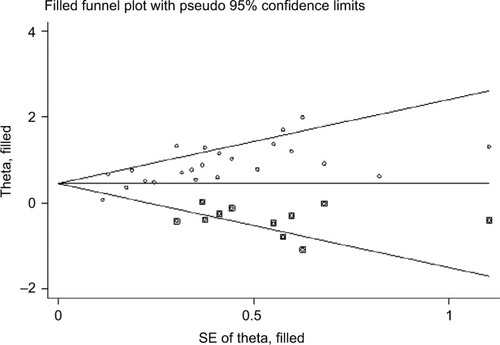
We would like to further explore some subgroups or more survival endpoints, such as Asian, lung cancer, disease-free survival, and the other you mentioned. But due to the lack of relevant researches, it cannot be achieved. So, more studies are needed to clarify these questions.
As mentioned by Dr Rama Jayaraj and Chellan Kumarasamy, in random-effects meta-analysis, it was more accurate to use τ2 parameter for estimating heterogeneity, when compared with χ2 and I2 values. Nevertheless, there is no difference between the two results in our study.
Based on the above, despite those limitations mentioned above, the final conclusion was still accurate. Overall, we really appreciate the comments from Dr Rama Jayaraj and Chellan Kumarasamy.
Disclosure
The authors declare no conflicts of interest in this communication.
References
- DaiCZhengYLiYPrognostic values of HE4 expression in patients with cancer: a meta-analysisCancer Manag Res2018104491450030349381
- ChengDSunYHeHThe diagnostic accuracy of HE4 in lung cancer: a meta-analysisDis Markers20152015617
- DayyaniFUhligSColsonBDiagnostic performance of risk of ovarian malignancy algorithm against CA125 and HE4 in connection with ovarian cancerInt J Gynecol Canc201626915861593
- MoherDLiberatiATetzlaffJAltmanDGPRISMA GroupPreferred reporting items for systematic reviews and meta-analyses: the PRISMA statementOpen Med200933e12313021603045
- DuvalSTweedieRA nonparametric “trim and fill” method of accounting for publication bias in meta-analysisJ Am Stat Assoc2000954498998
