Abstract
Background
Colorectal cancer (CRC) is among the most frequent and lethal malignancies worldwide. Although great advances have been made in the treatment of CRC, prognosis remains poor. Our previous study indicated that tripartite motif-containing 14 (TRIM14) was upregulated in CRC samples.
Methods
In the current study, the association between TRIM14 and CRC was investigated. Protein expression was determined by Western blotting and immunohistochemistry. Further, the biological roles of TRIM14 in CRC cell proliferation and apoptosis were explored both in vitro and in vivo.
Results
We observed that increased TRIM14 expression in CRC tissues was closely related with aggressive clinicopathological characteristics and poor prognosis. TRIM14 knockdown markedly reduced proliferation and increased apoptosis in HT-29 and SW620 cells, whereas TRIM14 overexpression in LoVo cells displayed opposite results. Xenograft experiments using HT-29 cells confirmed suppression of tumor growth and induction of apoptosis upon TRIM14 knockdown in vivo. Furthermore, downregulation of TRIM14 inhibited the AKT pathway, as indicated by reduced levels of phosphorylated AKT, Bcl-2 and Cyclin D1, and elevated levels of phosphatase and tensin homology (PTEN) and p27. In addition, TRIM14 colocalized with PTEN in the cytoplasm and induced PTEN ubiquitination. Moreover, PTEN overexpression significantly inhibited pro-proliferative effects of TRIM14, indicating an involvement of PTEN/AKT signaling in mediating TRIM14 functions.
Conclusions
The present data demonstrate that TRIM14 overexpression promotes CRC cell proliferation, suggesting TRIM14 as an attractive therapeutic target for CRC.
Keywords:
Introduction
Colorectal cancer (CRC) is a highly prevalent cancer in both males and females worldwide.Citation1 A number of risk factors have been associated with CRC development, including old age, obesity high fat intake, red meat consumption, smoking, and lack of physical exercise.Citation2–Citation4 Although great advances in screening and treatment methods have provided substantial benefits for patient outcomes, CRC remains the fourth deadliest cancer, causing approximately 700,000 deaths annually.Citation1 Therefore, broadening our understanding of CRC oncogenesis is critical in developing novel therapeutic targets for CRC.
Tripartite motif-containing proteins (TRIM) are a family of proteins that contain RING finger domain, B-box motif, and coiled-coil regionCitation5 with critical roles in regulating various biological processes, such as development, innate immune response, and cancer progression.Citation6,Citation7 A member of the TRIM family, TRIM14, was first found overexpressed in human immunodeficiency virus-associated human non-Hodgkin’s lymphomas.Citation8 Subsequent studies have shown that TRIM14 was involved in host defense against viral infections.Citation9,Citation10 Recent studies have revealed aberrant expression of TRIM14 in various human cancers. For instance, reduced expression of TRIM14 and functions in tumor suppression were observed in non-small cell lung cancer.Citation11 In contrast, oncogenic function and elevated expression of TRIM14 were reported in osteosarcoma,Citation12 oral squamous cell carcinoma,Citation13 tongue squamous cell carcinoma,Citation14 and hepatocellular carcinoma.Citation15
Activation of the phosphoinositide 3-kinase (PI3K)/AKT pathway, which triggers cell growth, proliferation, and motility,Citation16 has been related to CRC oncogenesis.Citation17 CRC cells overexpressing AKT displayed a highly proliferative and invasive state.Citation18 Phosphatase and tensin homology (PTEN), which antagonizes the effects of PI3K and ultimately inactivates the AKT pathway,Citation19 was found to be downregulated in CRCCitation20 and suppress CRC growth.Citation21 It has been reported that TRIM14 stimulates AKT signaling in osteosarcoma cells.Citation12 On the other hand, the association between TRIM14 and AKT signaling during CRC progression has not been explored.
The results of our recent studyCitation22 indicated that TRIM14 was upregulated in CRC and promoted the migration and invasion of CRC cells. In the current study, we further probed the correlation between TRIM14 expression and CRC patient prognosis, and went on to investigate the functions of TRIM14 on CRC cell proliferation and apoptosis. Moreover, we explored the involvement of PTEN/AKT signaling during this process.
Materials and methods
Patient information
The study was approved by the Ethics Committee at Yiwu Hospital, Wenzhou Medical University (Yiwu, China). Formalin-fixed, paraffin-embedded CRC samples, and matched non-cancerous tissue samples (n=74) were obtained from patients who received curative surgery at the Department of Gastroenterology, Yiwu Hospital (Yiwu, China) between 2009 and 2010 after written informed consent was obtained from every participant. Clinical information was retrieved from patient records.
Immunohistochemistry (IHC) analysis
Paraffin-embedded tissues were cut into 5-μm thickness sections, which were de-paraffinized, rehydrated, and subjected to IHC analysis with anti-TRIM14 antibody (Abcam, Cambridge, MA, USA; ab185349) as previously described. Twelve non-cancerous tissue samples were also stained as controls. IHC assessment was conducted by two investigators independently. The staining index was evaluated as follows: staining index = staining intensity (SI) × percentage of positive cells (PP). The SI was determined as 0, negative; 1, weak; 2, moderate; 3, strong. PP was classified as 0, <5%; 1, 5–25%; 2, 25–50%; 3, 50–75%; 4, >75%. Patients were classified into two groups (TRIM14 low expression and TRIM14 high expression) based on the staining index. The cut-off was set at 3.
Cell culture
Human CRC cell lines LoVo, HT-29, and SW620 (Shanghai Institute of Biochemistry and Cell Biology, Shanghai, China) were cultured in RPMI-1640 media containing 10% fetal bovine serum (Hyclone, Rockford, IL, USA) and antibiotics and maintained at 37°C and 5% CO2.
RNA interference-mediated knockdown of TRIM14 and overexpression of TRIM14 or PTEN
Lentiviral plasmids expressing control short hairpin RNA (shRNA) (shNC) or TRIM14 shRNAs (shTRIM14-1, 2, 3), and TRIM14 or PTEN complementary DNAs (cDNAs) were constructed, and lentiviruses were produced as previously described.Citation22
Western blot analysis
Cells were extracted in RIPA buffer with protease inhibitor cocktail (Beyotime, Shanghai, China). Protein was resolved in 10% or 15% SDS-PAGE gel, and Western blot analysis was performed according to standard protocol with GAPDH as a loading control. Primary antibodies included: TRIM14 (ab185349) from Abcam; PTEN (#9552), p-AKT (#9271), AKT (#9272), CyclinD1 (#2922), p27 and GAPDH (#5174) from Cell Signaling Technology (Danvers, MA, USA); and Bcl-2 (sc-130307) from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The band intensity was quantified by using ImageJ software (http://rsb.info.nih.gov/ij/, Bethesda, MD, USA).
Cell Counting Kit-8 (CCK-8) proliferation and apoptosis assays
(CCK-8 assay was performed to assess cell proliferation. Each cell line was plated into a 96-well tissue culture plate (2×103 cells per well). Following incubation for 0, 24, 48 , and 72 hrs at 37°C, cells were incubated with medium containing CCK-8 reagent (SAB biotech, College Park, MD, USA) at 37°C for 1 hr. At the end of incubation, optical density at 450 nm was measured by using a microplate reader (Labsystems, Helsinki, Finland). The well without cells served as blank control.
Cell apoptosis was evaluated using Annexin V assay. Cells were harvested, washed twice with ice-cold PBS and stained with Annexin V-fluorescein isothiocyanate and propidium iodide solution (Beyotime) in accordance with the manufacturer’s protocol. Cell apoptosis was measured by flow cytometry (Accuri C6, BD Biosciences, Franklin Lakes, NJ, USA), and data were analyzed using Accuri C6 software. Cells without staining were used as negative controls.
Tumor xenograft model
All animal experiments were approved by the Animal Care Committee of Yiwu Hospital, Wenzhou Medical University (Yiwu, China) and performed in accordance with procedures approved by the Animal Care Committee. HT-29 cells stably expressing shTRIM14 or shNC were established following viral transduction and selection with puromycin (Sigma) for 5 days. Cells (2×106 cells per mouse) were injected into the flank of four-week-old BALB/C nude mice (Shanghai Experimental Animal Center, n=6 per group). After tumors formed, length (L) and width (W) of tumors were measured with calipers every 3 days. Tumor volumes were calculated using the following formula: volume= (L×W2)/2. Mice were sacrificed on Day 33, and xenograft tumors were harvested, weighed, photographed, and paraffin-embedded. Five-μm thickness sections were cut, and IHC analysis with anti-Ki67 antibody (Abcam; ab15580) was performed to measure the proliferation index.
Real-time PCR analysis
Cells were lysed with Trizol reagent (Invitrogen Carlsbad, CA, USA) to isolate total RNA as per the manufacturer’s protocol. The concentration and quality of RNA were measured by NanoDrop spectrophotometer (ND-1000, Thermo Scientific, Rockford, IL, USA). Subsequently, complementary DNA (cDNA) was synthesized using total RNA and cDNA synthesis kit (Thermo Fisher), and then used as a template for real-time PCR analysis. Real-time PCR analysis was carried out using SYBR Green qPCR Master Mixes (Thermo Fisher) and ABI 7300 system (Applied Biosystem, Foster City, CA, USA) with GAPDH as an internal control. The oligonucleotides used as PCR primers were: PTEN, 5ʹ- TCAGGCGAGGGAGATGAGAG −3ʹ and 5ʹ- CAGGAGAAGCCGAGGAAGAG −3ʹ; Bcl-2, 5ʹ- GCAGTGTGGTCTCCGAATGTC −3ʹ and 5ʹ- CATTGCCTCTCCTCACGTTCC −3ʹ; Cyclin D1, 5ʹ- GCTGCGAAGTGGAAACCATC −3ʹ and 5ʹ- CCTCCTTCTGCACACATTTGAA −3ʹ; p27, 5ʹ- GCTTGCCCGAGTTCTACTAC −3ʹ and 5ʹ- GCAGGTCGCTTCCTTATTCC −3ʹ; GAPDH, 5ʹ-CACCCACTCCTCCACCTTTG-3ʹ and 5ʹ- CCACCACCCTGTTGCTGTAG −3ʹ.
Immunofluorescence staining
After washing with PBS, HT-29 cells cultured on coverslips were fixed with 4% paraformaldehyde at room temperature for 30 mins. To permeabilize cells and block non-specific protein–protein interactions, cells were incubated with 1% BSA and 10% normal goat serum in PBS-0.1% Tween for 1 hr. After probing with anti-PTEN (Abcam; ab79156; dilution: 1:100) and anti-TRIM14 (Invitrogen; PA5-62761; dilution: 1:200) antibodies overnight at 4°C, cells were labeled with Alexa Fluor 488 goat anti-rabbit IgG (H+L) (Beyotime; A0423; dilution: 1:500) and Alexa Fluor 555 donkey anti-mouse IgG (H+L) (Beyotime; A0460; dilution: 1:500) antibodies for 1 h at room temperature. Nuclei were then stained with 4, 6-diamino-2-phenylindole (DAPI; Beyotime), and cells were visualized with a fluorescence microscope.
Immunoprecipitation
Cells were lysed with RIPA buffer as described above. The supernatant was incubated with anti-PTEN (Cell Signaling Technology; #9552) or control IgG antibodies at 4°C for 2 hrs, followed by incubation with protein A agarose beads (Roche) at 4°C for another 1 hr. After washing with RIPA buffer three times, the protein was subjected to Western blotting analysis with anti-ubiquitin (Abcam; Ab7780) antibody.
Statistical analysis
The relationship between TRIM14 expression and clinicopathological features, and prognostic variables were analyzed by Fisher’s exact test and log-rank test, respectively. ANOVA and Student’s t-test were used to evaluate in vitro and in vivo experiments, respectively. Data are presented as the mean ± standard deviation (SD). Statistical analyses were performed using GraphPad Prism software (La Jolla, CA, USA). P-value less than 0.05 was considered statistically significant.
Results
Increased TRIM14 expression was correlated with poor CRC patient prognosis
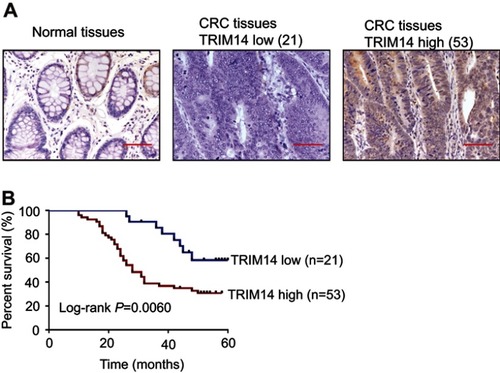
To determine the association between TRIM14 and clinicopathological characteristics, 74 CRC tissues were subjected to IHC staining with an antibody against TRIM14. Of the 74 CRC tissues, 53 (71.6%) cases with more than 25% of the positive-stained cells were defined as TRIM14 high expression (). As shown in , TRIM14 expression was significantly correlated with tumor size (P=0.0023) and clinical stage (P=0.0377), while no significant association between TRIM14 expression and other parameters, such as age, gender, and metastasis was observed. According to Kaplan–Meier analysis and log-rank test, CRC patients with high TRIM14 expression had shorter overall survival time (P=0.0060), as shown in . These data highlight a relationship between TRIM14 expression and CRC patient prognosis.
Table 1 Clinicopathological features and expression of TRIM14 in 74 colorectal cancer patients
Figure 1 Protein expression of TRIM14 in colorectal cancer (CRC). (A) TRIM14 protein expression in 74 CRC tissues were assessed byimmunohistochemistry (IHC) analysis. Noncancerous tissue samples were used as control. Scale bar: 50 μm; (B) Kaplan–Meier curves for overall survival of patients with CRC categorized according to TRIM14 expression.
TRIM14 expression affected proliferation of CRC cells
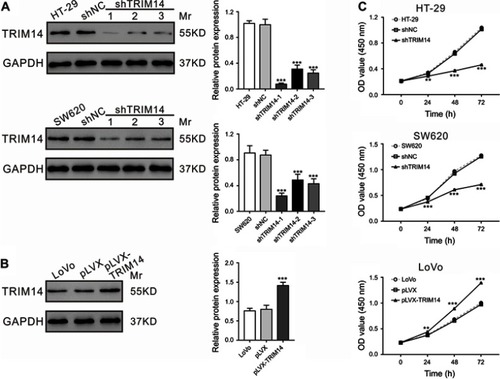
Given that TRIM14 expression was significantly correlated with tumor size, we then explored the potential roles of TRIM14 in CRC cell proliferation by knocking down TRIM14 in HT-29 and SW620 cells using TRIM14 shRNA, or by overexpressing TRIM14 in LoVo cells by lentiviral infection as previously described.Citation22 TRIM14 protein expression was assessed by Western blotting at 48 hrs post-lentiviral infection. As shown in , TRIM14 protein levels were nearly abolished in cells with shTRIM14-1. Hence, this shRNA was used in the subsequent experiments. Similarly, LoVo cells with pLVX-TRIM14 infection displayed elevated protein levels of TRIM14, as shown in . To determine CRC cell proliferation, we performed CCK-8 assays at 24, 48, and 72 hrs post-viral infection. Compared to the control, HT-29 and SW620 cells with shTRIM14 displayed remarkably lower proliferation. On the other hand, LoVo cells with pLVX-TRIM14 infection showed notably higher proliferation (). Together, these data demonstrate a pro-proliferative function of TRIM14 in CRC.
Figure 2 TRIM14 expression affected the proliferation of colorectal cancer (CRC) cells. (A) TRIM14 expression in HT-29 and SW620 cells after infection with shTRIM14 or shNC lentivirus. (B) TRIM14 expression in LoVo cells after infection with pLVX-TRIM14 or pLVX lentivirus. (C) CCK-8 assays were carried out to assess the proliferation of CRC cells infected with shNC, shTRIM14, pLVX, and pLVX-TRIM14, respectively. shNC or pLVX served as negative control. **P<0.01, ***P<0.001 vs shNC or pLVX. Abbreviation: shNC, lentiviral plasmid expressing control short hairpin RNA.
TRIM14 regulated apoptosis of CRC cells
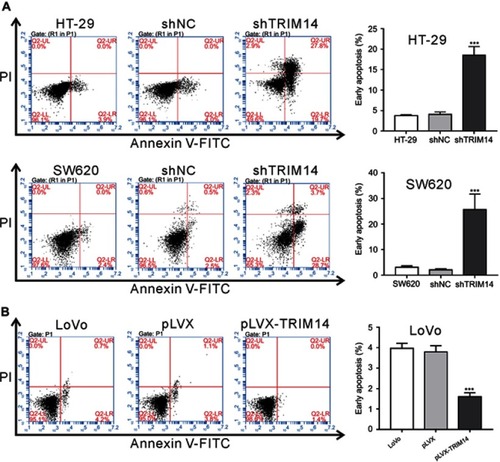
To elucidate the role of TRIM14 in cell apoptosis, Annexin V assay and flow cytometry analysis were performed. The results showed that early apoptotic rates were significantly increased in HT-29 and SW620 cells with shTRIM14 compared to control cells (), whereas opposite effects were seen in LoVo cells with pLVX-TRIM14 infection (). Hence, our results indicate an inhibitory effect of TRIM14 on CRC cell apoptosis.
Figure 3 TRIM14 regulated the apoptosis of colorectal cancer (CRC) cells. (A) HT-29 and SW620 cells were infected with shTRIM14 or shNC lentivirus. (B) LoVo cells were infected with pLVX-TRIM14 or pLVX lentivirus. At 48 hrs after infection, flow cytometry analysis was used to analyze apoptotic rates of CRC cells. shNC or pLVX served as negative control. ***P<0.001 vs shNC or pLVX.
Abbreviation: shNC, lentiviral plasmid expressing control short hairpin RNA.
Knockdown of TRIM14 affected cell proliferation and apoptosis of CRC cells in vivo
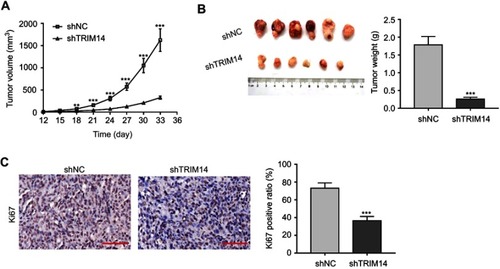
To further investigate the effects of TRIM14 knockdown on CRC tumor growth in vivo, HT-29 cells stably expressing shTRIM14 or shNC were subcutaneously injected into nude mice. As shown in , xenograft tumors generated from cells with shTRIM14 displayed slower growth than those formed in cells with shNC. On Day 33, tumor volume was significantly lower in the shTRIM14 group than in shNC group (). IHC () analysis also revealed a low percentage of Ki67-positive cells in the shTRIM14 group. Together, these data indicate that TRIM14 may serve as an oncogene in CRC in vivo.
Figure 4 Knockdown of TRIM14 affected proliferation and apoptosis of colorectal cancer (CRC) cells in vivo. HT-29 cells stably expressing shTRIM14 or shNC were injected subcutaneously into nude mice. (A) Tumor volumes were measured every three days, and presented as growth curves. (B) On Day 33, xenograft tumors were recovered, weighed, and photographed. (C) Tumor sections were subjected to IHC staining using anti-Ki67 antibody. Scale bar: 50 μm. shNC served as negative control. **P<0.01, ***P<0.001 vs shNC.
Abbreviation: shNC, lentiviral plasmid expressing control short hairpin RNA.
TRIM14 regulated PTEN/AKT signaling
The AKT pathway is critical for the oncogenesis of CRC.Citation17 Given the report that TRIM14 overexpression in osteosarcoma cells activated the AKT signaling pathway,Citation12 we examined the protein levels of PTEN, p-AKT AKT, and downstream effectors of the AKT pathway (BCl-2,Citation23 Cyclin D1,Citation24 and p27Citation25) in CRC cells with TRIM14 knockdown by Western blotting. Interestingly, TRIM14 knockdown suppressed the levels of p-AKT, Bcl-2, and Cyclin D1, but enhanced the levels of PTEN and p27 (). On the contrary, TRIM14 overexpression displayed the reverse effects ().
Figure 5 TRIM14 regulated the PTEN/AKT signaling pathway. (A, B) Protein levels of PTEN, p-AKT, AKT, Bcl-2, Cyclin D1, and p27 were determined in the indicated colorectal cancer (CRC) cells at 48 hrs after viral infection. (C, D) mRNA expression of PTEN, Bcl-2, Cyclin D1, and p27 was assessed in the indicated CRC cells at 48 hrs after viral infection. shNC or pLVX served as negative control. ***P<0.001 vs shNC or pLVX. (E) LoVo cells were fixed and stained with TRIM14 (green) and PTEN (red) antibodies. Nuclei were stained with DAPI (blue). Representative images are shown. Scale bar: 20 μm. (F) Effects of TRIM14 on PTEN ubiquitination.
Abbreviation: shNC, lentiviral plasmid expressing control short hairpin RNA.
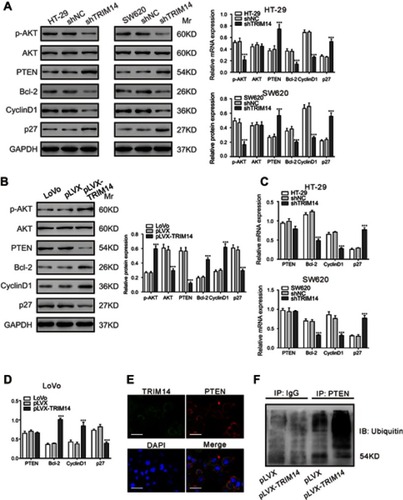
Real-time PCR analysis showed that TRIM14 knockdown had similar effects on the mRNA expression of Bcl-2, Cyclin D1, and p27. Notably, we observed that the PTEN mRNA was not affected by TRIM14 overexpression or knockdown ( and ). As most TRIM family proteins have E3 ubiquitin ligase activity,Citation7 we speculated that TRIM14 may increase PTEN ubiquitination. Using TRIM14 and PTEN antibodies, we performed immunofluorescence staining in LoVo cells. As shown in , TRIM14 and PTEN localized in the cytoplasm and membrane, suggesting that TRIM14 may form a complex with PTEN. PTEN ubiquitination was also assessed in LoVo cells that were infected with pLVX or pLVX-TRIM14. As shown in , PTEN ubiquitination was increased by TRIM14 overexpression. Collectively, these data suggest that TRIM14 regulates PTEN expression via post-translational modification.
Overexpression of PTEN suppressed TRIM14-mediated CRC cell proliferation
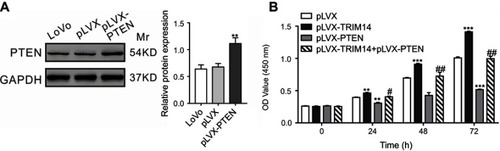
To determine whether PTEN/AKT was involved in TRIM14-induced proliferation of CRC cells, we examined the growth ability of LoVo cells overexpressing TRIM14 and PTEN. As indicated in , PTEN overexpression inhibited the pro-proliferative effects of TRIM14. Hence, these data demonstrate that PTEN/AKT pathway contributes to the functions of TRIM14 in promoting CRC cell proliferation.
Figure 6 Overexpression of PTEN suppressed TRIM14-mediated colorectal cancer (CRC) cell proliferation. (A) PTEN was highly expressed in LoVo cells after infection with pLVX-PTEN virus. (B) CCK-8 assays were carried out to assess the proliferation of LoVo cells with PTEN and TRIM14 overexpression. **P<0.01, ***P<0.001 vs pLVX; #P<0.05, ##P<0.01 vs pLVX-TRIM14.
Discussion
The TRIM protein family is known to contain more than 70 members and most of them participate in numerous biological processes, such as innate immune response, cell proliferation, apoptosis, and invasion.Citation6,Citation7 TRIM14, a member of this family, is induced in many cell types, which is related to the modulation of diverse cellular functions,Citation8,Citation12–Citation15,Citation26 including cancer progression. The upregulation and oncogenic functions of other TRIM proteins, such as TRIM24Citation27,Citation28 and TRIM29Citation29,Citation30 have been reported in CRC. Recently, we have reported that TRIM14 mRNA levels were elevated in CRC tissues.Citation22 In the current study, IHC analysis confirmed the above findings at the translational level. Further, our data suggested that elevated expression of TRIM14 in CRC tissues was closely related to the aggressive clinicopathological characteristics and poor patient prognosis (). TRIM14 knockdown inhibited CRC cell proliferation and promoted apoptosis, while TRIM14 overexpression had the opposite effects (–). These data, combined with our previous study,Citation22 demonstrated the oncogenic effects of TRIM14 in CRC, which were consistent with the findings in other types of human malignancies.Citation12–Citation15
We also explored the possible mechanism through which TRIM14 induced proliferation and inhibited apoptosis of CRC cells. It has been well established that AKT signaling is critical in the tumorigenesis and development of cancer by regulating cell proliferation, apoptosis, and migration.Citation16 AKT phosphorylation predicts poor disease-free survival in stage II CRC patients.Citation31 CRC cells with AKT overexpression were highly proliferative and invasive.Citation18 The blockage of the AKT pathway possesses anti-tumorigenic effects in CRC cells and provides a potential strategy for CRC treatment.Citation32–Citation34 As a negative regulator of the AKT pathway, PTEN is downregulated in CRCCitation20 and serves as a tumor suppressor in CRC.Citation21 Herein, consistent with earlier findings, TRIM14 overexpression increased p-AKT levels and decreased PTEN levels in CRC cells (). The AKT pathway exerts its functions by regulating multiple downstream molecules, such as Cyclin D1, p27, and Bcl-2. Cyclin D1 is an important factor responsible for G1/S transition,Citation24 and p27 is an inhibitor of Cyclin-dependent kinase,Citation25 while Bcl-2 is a well-known anti-apoptotic factor.Citation23 In the present study, TRIM14 knockdown resulted in reduced Cyclin D1 and Bcl-2 levels but enhanced p27 levels in CRC cells. Opposite effects were seen in CRC cells with TRIM14 overexpression (). Notably, overexpression of PTEN reversed the pro-proliferative effects of TRIM14 on CRC cells (). Accordingly, these data indicate that TRIM14 overexpression activates the AKT pathway, which contributes to the highly proliferative phenotype of CRC cells.
Lastly, it has been reported that polyubiquitination of PTEN caused its retention in the cytoplasm and degradation.Citation35 TRIM27 is able to promote PTEN polyubiquitination and thus inhibits its phosphatase activity.Citation36 In the current study, TRIM14 levels affected PTEN expression at translational level, but not at the transcriptional level ( and ). Further, we found that TRIM14 may form a complex with PTEN and promoted PTEN ubiquitination ( and ), which was similar to that observed for TRIM27. However, TRIM14 lacks a RING domain, which is usually required for E3 ubiquitin ligase activity on TRIM family proteins.Citation7 The precise mechanism of how TRIM14 affected PTEN ubiquitination requires further investigation.
In summary, we reported that elevated expression of TRIM14 in clinical CRC samples was correlated with patient prognosis. Knockdown of TRIM14 suppressed the aggressiveness of CRC cells by modulating the PTEN/AKT signaling pathway. TRIM14 formed a complex with PTEN and induced PTEN ubiquitination. These findings advance our knowledge of CRC progression and suggest that TRIM14 may emerge as an attractive therapeutic target for CRC.
Disclosure
The authors declare that they have no competing or conflicting interests in regard to this work.
Acknowledgment
This study was supported by a grant from the Zhejiang Science and Technology Project (2017C33179).
References
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.2126225651787
- Kuiper JG, Phipps AI, Neuhouser ML, et al. Recreational physical activity, body mass index, and survival in women with colorectal cancer. Cancer Causes Control. 2012;23(12):1939–1948. doi:10.1007/s10552-012-0071-223053793
- Slattery ML. Diet, lifestyle, and colon cancer. Semin Gastrointest Dis. 2000;11(3):142–146.10950460
- Huxley RR, Ansary-Moghaddam A, Clifton P, Czernichow S, Parr CL, Woodward M. The impact of dietary and lifestyle risk factors on risk of colorectal cancer: a quantitative overview of the epidemiological evidence. Int J Cancer. 2009;125(1):171–180. doi:10.1002/ijc.2434319350627
- Meroni G, Diez‐Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’E3 ubiquitin ligases. Bioessays. 2005;27(11):1147–1157. doi:10.1002/bies.2030416237670
- Ozato K, Shin D-M, Chang T-H, Morse HC III. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8(11):849–860. doi:10.1038/nri241318836477
- Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11(11):792–804. doi:10.1038/nrc313921979307
- Tarantul V, Nikolaev A, Hannig H, et al. Detection of abundantly transcribed genes and gene translocation in human immunodeficiency virus-associated non-Hodgkin’s lymphoma. Neoplasia. 2001;3(2):132–142. doi:10.1038/sj/neo/790013711420749
- Tan P, He L, Cui J, et al. Assembly of the WHIP-TRIM14-PPP6C mitochondrial complex promotes RIG-I-mediated antiviral signaling. Mol Cell. 2017; 68(2):293–307. e295. doi:10.1016/j.molcel.2017.09.03529053956
- Wang S, Chen Y, Li C, et al. TRIM14 inhibits hepatitis C virus infection by SPRY domain-dependent targeted degradation of the viral NS5A protein. Sci Rep. 2016;6:32336. doi:10.1038/srep3233627578425
- Hai J, Zhu CQ, Wang T, Organ SL, Shepherd FA, Tsao MS. TRIM14 is a putative tumor suppressor and regulator of innate immune response in non-small cell lung cancer. Sci Rep. 2017;7:39692. doi:10.1038/srep3969228059079
- Xu G, Guo Y, Xu D, et al. TRIM14 regulates cell proliferation and invasion in osteosarcoma via promotion of the AKT signaling pathway. Sci Rep. 2017;7:42411. doi:10.1038/srep4241128205534
- Wang T, Ren Y, Liu R, et al. miR-195-5p suppresses the proliferation, migration, and invasion of oral squamous cell carcinoma by targeting TRIM14. Biomed Res Int. 2017;2017:1–13. doi:10.1155/2017/6490349
- Su X, Wang J, Chen W, Li Z, Fu X, Yang A. Overexpression of TRIM14 promotes tongue squamous cell carcinoma aggressiveness by activating the NF-kappaB signaling pathway. Oncotarget. 2016;7(9):9939–9950. doi:10.18632/oncotarget.694126799420
- Dong B, Zhang W. High levels of TRIM14 are associated with poor prognosis in hepatocellular carcinoma. Oncol Res Treat. 2018;41(3):129–134. doi:10.1159/00048562529485416
- Mayer IA, Arteaga CL. The PI3K/AKT pathway as a target for cancer treatment. Annu Rev Med. 2016;67:11–28. doi:10.1146/annurev-med-062913-05134326473415
- Huang XF, Chen JZ. Obesity, the PI3K/Akt signal pathway and colon cancer. Obes Rev. 2009;10(6):610–616. doi:10.1111/j.1467-789X.2009.00607.x19527447
- Suman S, Kurisetty V, Das TP, et al. Activation of AKT signaling promotes epithelial–mesenchymal transition and tumor growth in colorectal cancer cells. Mol Carcinog. 2014;53:S1. doi:10.1002/mc.22076
- Martini M, De Santis MC, Braccini L, Gulluni F, Hirsch E. PI3K/AKT signaling pathway and cancer: an updated review. Ann Med. 2014;46(6):372–383. doi:10.3109/07853890.2014.91283624897931
- Colakoglu T, Yildirim S, Kayaselcuk F, et al. Clinicopathological significance of PTEN loss and the phosphoinositide 3-kinase/Akt pathway in sporadic colorectal neoplasms: is PTEN loss predictor of local recurrence? Am J Surg. 2008;195(6):719–725. doi:10.1016/j.amjsurg.2007.05.06118440486
- Sun Y, Tian H, Wang L. Effects of PTEN on the proliferation and apoptosis of colorectal cancer cells via the phosphoinositol-3-kinase/Akt pathway. Oncol Rep. 2015;33(4):1828–1836. doi:10.3892/or.2015.380425683168
- Jin Z, Li H, Hong X, et al. TRIM14 promotes colorectal cancer cell migration and invasion through the SPHK1/STAT3 pathway. Cancer Cell Int. 2018;18(1):202.30555277
- Pugazhenthi S, Nesterova A, Sable C, et al. Akt/protein kinase B up-regulates Bcl-2 expression through cAMP-response element-binding protein. J Biol Chem. 2000;275(15):10761–10766. doi:10.1074/jbc.275.15.1076110753867
- Fatrai S, Elghazi L, Balcazar N, et al. Akt induces β-cell proliferation by regulating cyclin D1, cyclin D2, and p21 levels and cyclin-dependent kinase-4 activity. Diabetes. 2006;55(2):318–325. doi:10.2337/diabetes.55.02.06.db05-075716443763
- Sun H, Lesche R, Li D-M, et al. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3, 4, 5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc Natl Acad Sci U S A. 1999;96(11):6199–6204. doi:10.1073/pnas.96.11.619910339565
- Zhou Z, Jia X, Xue Q, et al. TRIM14 is a mitochondrial adaptor that facilitates retinoic acid-inducible gene-I-like receptor-mediated innate immune response. Proc Natl Acad Sci U S A. 2014;111(2):E245–E254. doi:10.1073/pnas.131694111124379373
- Wang J, Zhu J, Dong M, Yu H, Dai X, Li K. Knockdown of tripartite motif containing 24 by lentivirus suppresses cell growth and induces apoptosis in human colorectal cancer cells. Oncol Res. 2014;22(1):39–45. doi:10.3727/096504014X1407843600501225700357
- Wang F-Q, Han Y, Yao W, Yu J. Prognostic relevance of tripartite motif containing 24 expression in colorectal cancer. Pathol Res Pract. 2017;213(10):1271–1275. doi:10.1016/j.prp.2017.08.00828916426
- Jiang T, Tang HM, Lu S, Yan DW, Yang YX, Peng ZH. Up-regulation of tripartite motif-containing 29 promotes cancer cell proliferation and predicts poor survival in colorectal cancer. Med Oncol. 2013;30(4):715. doi:10.1007/s12032-013-0715-424078150
- Xu W, Xu B, Yao Y, et al. RNA interference against TRIM29 inhibits migration and invasion of colorectal cancer cells. Oncol Rep. 2016;36(3):1411–1418. doi:10.3892/or.2016.494127430345
- Malinowsky K, Nitsche U, Janssen K, et al. Activation of the PI3K/AKT pathway correlates with prognosis in stage II colon cancer. Br J Cancer. 2014;110(8):2081–2089. doi:10.1038/bjc.2014.10024619078
- Dong M, Yang G, Liu H, et al. Aged black garlic extract inhibits HT29 colon cancer cell growth via the PI3K/Akt signaling pathway. Biomed Rep. 2014;2(2):250–254. doi:10.3892/br.2014.22624649105
- Li XX, Huang LY, Peng JJ, et al. Klotho suppresses growth and invasion of colon cancer cells through inhibition of IGF1R-mediated PI3K/AKT pathway. Int J Oncol. 2014;45(2):611–618. doi:10.3892/ijo.2014.243024818842
- Enayat S, Mş C, Başaran AA, Gürsel M, Banerjee S. Anticarcinogenic effects of the ethanolic extract of Salix aegyptiaca in colon cancer cells: involvement of Akt/PKB and MAPK pathways. Nutr Cancer. 2013;65(7):1045–1058. doi:10.1080/01635581.2013.85096624168160
- Trotman LC, Wang X, Alimonti A, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128(1):141–156. doi:10.1016/j.cell.2006.11.04017218261
- Lee JT, Shan J, Zhong J, et al. RFP-mediated ubiquitination of PTEN modulates its effect on AKT activation. Cell Res. 2013;23(4):552–564. doi:10.1038/cr.2013.2723419514
