Abstract
Objective
Recent studies have shown that circRNAs participate in ovarian cancer progression and act as potential biomarkers for ovarian cancer diagnosis and prognosis. In the present study, we aimed to investigate the expression pattern and prognostic significance of circ_0078607 in high-grade serous ovarian cancer (HGSOC).
Methods
The expression of circ_0078607 was detected by quantitative real-time polymerase chain reaction (qRT-PCR) in 49 cases of HGSOC. Clinical data of patients with HGSOC were retrospectively collected, and those patients were divided according to their expression of circ_0078607. Correlation between circ_0078607 and clinical features as well as the prognosis in patients with HGSOC was analyzed. t-test and chi-square test were used to compare continuous and categorical variables. The Cox hazard regression model was used to assess prognostic factors. Both progression-free survival (PFS) and overall survival (OS) curves were generated by Kaplan–Meier method.
Results
The expression of circ_0078607 was significantly downregulated in ovarian cancer tissues compared with adjacent non-cancerous tissues. Besides, patients with low circ_0078607 expression exhibited parameters associated with poor prognosis, including advanced FIGO stage and higher serum CA125 level. Kaplan–Meier survival curve analysis showed that patients with low circ_0078607 expression had shorter PFS and OS. Cox regression analysis showed that low expression of circ_0078607 was a predictor for poor PFS and OS in HGSOC patients.
Conclusion
Low expression of circ_0078607 might be an adverse prognostic indicator for HGSOC.
Keywords:
Introduction
Ovarian cancer is the most lethal gynecological cancer globally.Citation1 High-grade serous ovarian cancer (HGSOC) is the most common subtype (70%) associated with a higher degree of malignancy and a poor prognosis.Citation2 Throughout the last three decades, major improvements have been made in the fields of surgical debulking as well as chemotherapy and targeted therapy, the long-term survival of ovarian cancer patients remains unsatisfactory, with few effective prognostic biomarkers available at present.Citation2,Citation3 In most cases, ovarian cancer is usually detected in advanced stages.Citation4 Therefore, there is an urgent need for new molecular markers to predict the prognosis of ovarian cancer patients to guide clinical treatment.
As a naturally occurring family of long noncoding RNAs (lncRNAs), circRNAs are covalently closed, single-stranded circular transcripts with no 5′ caps or 3′ polyA tails, this structural characteristic makes circRNAs stable, conserved, highly abundant, and dynamically expressed in a special manner in a variety of tissues.Citation5,Citation6 With the rapid development of high-throughput sequencing, RNA sequencing technology, and bioinformatics, the abundance and diversity of circRNAs were gradually revealed.Citation7 Numerous studies demonstrated that circRNAs played an important regulatory role in the progression of various types of human tumors, including bladder cancer,Citation8 breast cancer,Citation9 and colorectal cancer.Citation10 In addition, the function of circRNAs observed in these cancers was also shown in ovarian cancer. Some studies provided evidence that circRNAs were differentially expressed in tumor tissue of ovarian cancer and played an important role in carcinogenesis because of participating in multiple cancer-related pathways, including cell proliferation, migration, invasion, and apoptosis.Citation11–Citation13 Due to their expression patterns and multiple regulating functions in ovarian cancer tissues, circRNAs showed great potential as prognostic biomarkers in ovarian cancer.Citation11,Citation14,Citation15
Our previous studies had shown that circ_0078607 was low expressed in ovarian cancer tissue compared with adjacent non-cancerous tissue.Citation16 However, it is still unclear whether circ_0078607 could serve as a potential biomarker of ovarian cancer. So, in this study, we performed correlation analysis to investigate the association between circ_0078607 expression and clinical features and prognosis in HGSOC patients.
Methods
Patients and Specimens
This project was approved by the Ethics Committee of Renji Hospital, School of Medicine, Shanghai Jiaotong University, and informed consent in compliance with the declaration of Helsinki obtained from each patient. To investigate a histopathological homogenous group, only HGSOC was included, and the sample had been all pathologically confirmed by pathologists. A total of 49 pairs of HGSOC and corresponding adjacent non-cancerous tissues were collected from patients who had received surgery at the Department of Gynecology in Renji hospital. None of the patients had previous cases of cancer or other serious organ system diseases. None of the enrolled patients had received chemotherapy or radiotherapy prior to surgery and the neoplasm staging was in accordance with the International Federation of Gynecology and Obstetrics (FIGO). All tissues were immersed in liquid nitrogen and preserved at −80°C until use. All the patients got follow-up for 38 months from the date of surgical resection. Progression-free survival (PFS) was defined from the initial surgery to disease progression. Overall survival (OS) was defined from the initial surgery to patient death from any cause or the last follow-up.
RNA Isolation and Quantification
Total RNA from tissues was extracted with TRIzol reagent. The RNA was reverse transcribed to cDNA following the instructions of reverse transcription kit PrimeScript® RT Master Mix Perfect Real Time kit (TaKaRa, Dalian, China). Real-time quantitative reverse transcription-polymerase chain reaction (RT-PCR) was performed using SYBR Green qPCR Master Mix (Yeasen) on an ABI PRISM 7500 fast Sequence Detection System (Applied Biosystems, Foster City, CA, USA) following the protocols. Primers were designed to perform the amplification, and the sequences were as follows: GAPDH, Forward: 5′-TATGATGATATCAAGAGGGTAGT-3′ and Reverse: 5′-TGTATCCAAACTCATTGTCATAC-3′; circ_0078607, Forward: 5′-CGGAATTCTGAAATATGCTATCTTACAGTTTGACCTTGTCTGTGTCAATG-3′ and Reverse: 5′-CGGGATCCTCAAGAAAAAATATATTCACCTCTGAGTAATTTGATGAGAGG-3′; GAPDH as an internal control. The comparative expression level was compared using 2−ΔΔCt method.
Statistical Analysis
Statistical analysis was performed using SPSS statistical software version 26.0 (SPSS Inc., Chicago, IL, USA) for Windows. Student’s t-test was used to compare quantitative data between groups. The Chi-square test was used to determine association of clinical features with circ_0078607. The cumulative PFS and OS rates were calculated using the Kaplan–Meier method, and differences were compared using a Log rank test. A Cox proportional hazard regression model was used for univariate and multivariate analyses. P<0.05 was considered statistically significant.
Results
Clinical Characteristics of HGSOC Patients
The age of patients ranged from 41–85 years (median 60 years). Three patients in FIGO stage I (6.12%), seven patients in FIGO stage II (14.28%), thirty-three patients in FIGO stage III (67.35%) and six patients in FIGO stage IV (12.25%) disease. The maximum diameter of the tumor was less than 10 cm in 36 (73.47%) patients and greater than 10 cm in 13 (26.53%) patients. Ascites volume was less than 1000 mL in 27 (55.10%) patients and greater than 1000 mL in 22 (44.90%) patients. Preoperative serum CA125 level was below 500ng/mL in 22 (44.89%) patients and above 500ng/mL in 27 (55.11%) patients. All of patients received comprehensive staging and tumor-cell surgical debulking followed by six cycles of platinum-based chemotherapy as follows: paclitaxel (175 mg/m2) and carboplatin (AUC5). Regarding sensitivity to chemotherapy, tumors recurring within 6 months were defined as platinum resistance, conversely, tumors recurring over 6 months were defined as platinum sensitive. 35 (71.43%) patients were classified as platinum-sensitive and 14 (28.57%) patients as platinum-resistant. All the patients got follow-up after surgical resection. We used the median of circ_0078607 expression as a cut-off to divide patients into a circ_0078607 low-expression group, which included 31 (63.27%) patients, and a circ_0078607 high-expression group, which included 18 (36.73%) patients. Detailed characteristic information of the patient cohort is summarized in .
Table 1 The Baseline Information of High-Grade Serous Ovarian Cancer Patients (HGSOC) (N=49)
Reduced Expression of circ_0078607 in HGSOC
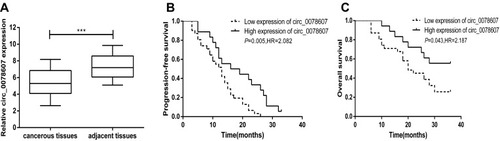
qRT-PCR analysis was performed to measure the expression of circ_0078607 in 49 pairs of HGSOC tissues and their corresponding adjacent non-cancerous tissues. The result revealed that lower circ_0078607 expression was found in HGSOC tissues compared with corresponding adjacent non-cancerous tissues (P<0.001, ).
Figure 1 Expression of circ_0078607 in HGSOC and its correlation with prognosis. (A) Relative expression of circ_0078607 in 49 pairs of HGSOC tissues and their corresponding adjacent non-cancerous tissues. (B) Correlation between circ_0078607 expression and PFS in HGSOC. (C) Correlation between circ_0078607 expression and OS in HGSOC. ***p < 0.001.
Correlation Between circ_0078607 Expression and Clinical Features in HGSOC Patients
To better understand the significance of circ_0078607 in HGSOC, the association between the expression of circ_0078607 and the clinical parameters of 49 patients was analyzed. As shown in , circ_0078607 expression showed no association with the clinical features of age (P=0.483), tumor size (P=0.603), or the amount of ascites (P=0.066). Besides, there was no statistically significant correlation between expression of circ_0078607 and platinum-based chemotherapy sensitivity. However, circ_0078607 expression was significantly correlated with advanced FIGO stage (P=0.014) and higher serum CA125 level (P=0.019). Among patients with low expression of circ_0078607, 90.3% of patients were in FIGO stage III–IV, and 61.7% of patients had preoperative serum CA125 level higher than 500ng/mL. While in patients with high expression of circ_0078607, 61.1% of patients were in FIGO stage III–IV. 33.3% of patients had preoperative serum CA125 level higher than 500ng/mL. In addition, patients with low expression of circ_0078607 had higher mortality rate. These data suggested that patients with low expression of circ_0078607 had a worse prognosis.
Table 2 Correlations Between circ_0078607 Expression and Clinical Features in HGSOC Patients
Association Between circ_0078607 Expression and the Prognosis in HGSOC Patients
To further assess the potential clinical value of circ_0078607 expression for the prognosis in ovarian cancer patients, we used Kaplan–Meier survival curve analysis and Log rank tests to evaluate the association between circ_0078607 expression and PFS and OS. In 49 HGSOC patients, a significant correlation was found between low circ_0078607 expression and PFS and OS. Both PFS and OS were significantly shortened in patients with low circ_0078607 expression ().
We then performed the univariate and multivariate Cox regression analysis for clinical factors in relation to PFS and OS of HGSOC patients. As shown in , in univariate Cox regression model analysis, low circ_0078607 expression (HR=2.083, P=0.005), advanced FIGO stage (HR=2.629, P=0.002), higher serum CA125 level (HR=2.025, P=0.007), and larger amount of ascites (HR=2.421, P=0.011) were all associated with shorter PFS, while multivariate Cox regression model suggested that low circ_0078607 expression (HR=2.180, P=0.024), advanced FIGO stage (HR=3.112, P= 0.011), higher serum CA125 level (HR=2.672, P=0.036) were risk factors for PFS of HGSOC patients.
Table 3 Univariate and Multivariate Cox Analysis of Progression-Free Survival (PFS) in HGSOC
As shown in , in univariate Cox regression model analysis, low circ_0078607 expression (HR=2.187, P=0.043) and advanced FIGO stage (HR=4.586, P=0.018) were shown to be significant for the overall survival of high-grade serous ovarian cancer patients. The subsequent multivariate Cox regression model analysis also revealed that low circ_0078607 expression (HR=1.795, P=0.038) and advanced FIGO stage (HR=2.149, P=0.028) emerged as predictors for poor overall survival, based on their association with shorter overall survival. These results provided evidence that low expression of circ_0078607 indicated a worse prognosis of patients with HGSOC.
Table 4 Univariate and Multivariate Cox Analysis of Overall Survival (OS) in HGSOC
Discussion
Ovarian cancer is a deadly disease owing to insidious onset, early metastasis and high rate of recurrence after standard of care.Citation17 HGSOC is the most common type of ovarian cancer and the prognosis is usually very poor. Although great progress has been made in the treatment of ovarian cancer, the OS of these patients has not been significantly improved, and the outcome of ovarian cancer remains difficult to predict.Citation18 Thus, novel biomarkers that can be used to predict the prognosis of ovarian cancer remain urgently needed.
Many circRNAs had been shown to be dysregulated in ovarian cancer tissues and had been proved to regulate the progression of ovarian cancer in a variety of ways.Citation19,Citation20 For example, circEPSTI1 was reported overexpressed in ovarian cancer, this circRNA was capable to sponge miR-942 to upregulate EPSTI1 to promote the progress of ovarian cancer.Citation19 Chen et al reported that circCDR1as, which was significantly lower in ovarian cancer tissues, could function as a tumor suppressor by sponging miR-135b-5p, which exerts a cancer-promoting effect by targeting HIF1AN in ovarian cancer.Citation21 Chen et al reported that circ_0061140 was upregulated in OC cell lines and positively regulates FOXM1 expression by acting as a competing endogenous RNA for miR-370 binding.Citation22 Our previous studies had shown that circ_0078607, which was down-regulated in ovarian cancer, could serve as a sponge of miR-518a-5p to elevate Fas expression to inhibit proliferation and invasion and promote apoptosis of ovarian cancer cells.Citation16 Some circRNAs had been reported to be potential diagnostic and prognosis biomarkers in ovarian cancer, and their deregulation was associated with clinicopathological features in ovarian cancer patients. For example, Chen et al reported that expression of circ-ABCB10 was correlated with poor differentiation and advanced FIGO stage in epithelial ovarian cancer.Citation23 Liu et al reported that high circHIPK3 expression was an independent predictor of OS in epithelial ovarian cancer patients.Citation14 Gao et al reported that expression of circRNA1656 could be expected to become a novel tumor marker for ovarian cancer.Citation24
We first identified hsa_circ_0078607, derived from exonic back-splicing of SLC22A3 gene, was downregulated in HGSOC tissues comparing with corresponding adjacent non-cancerous ovarian tissues. However, the relationship between circ_0078607 and the clinical features and prognosis of HGSOC had not been studied. The current study aimed to detect the association between circ_0078607 expression and the clinical features of patients with HGSOC and the predictive value of circ_0078607 in HGSOC. Our study showed that patients with low circ_0078607 expression exhibited parameters associated with poor prognosis, including advanced FIGO stage and higher serum CA125 level, indicating patients with low circ_0078607 expression may present a poor prognosis. In addition, patients with low circ_0078607 expression were more likely to die. However, the expression of circ_0078607 was not significantly correlated with age, tumor size, ascites, and chemosensitivity.
We used Kaplan–Meier survival curve to evaluate the effects of circ_0078607 expression on clinical prognosis of HGSOC patients, and the results revealed low circ_0078607 expression was associated with shorter PFS and OS. In 31 patients with low circ_0078607 expression, the median PFS time was 13 months and the 3-year survival rate was 25.81%, in 18 patients with high circ_0078607 expression, the median PFS time was 17.5 months and the 3-year survival rate was 55.56%. The subsequent Cox regression model analysis showed that low expression of circ_0078607 could serve as a powerful predictor of the poor prognosis for patients with HGSOC.
The disadvantage of this study is that the sample size of our study was small and the follow-up time was not long enough, but we continue expanding the sample size to verify the efficacy of circ_0078607 as diagnostic and prognostic biomarkers for ovarian cancer. Meanwhile, follow-ups are proceeded with patients who are alive. Besides, future research can not only focus on the role of circ_0078607 in the overall management of ovarian cancer patients, but also monitor the expression of circ_0078607 at different periods. To explore the role of this marker in prospectively tracking and predicting the progression and prognosis of patients with ovarian cancer.
Conclusions
Our study demonstrated the significant prognosis importance of circ_0078607 in HGSOC patients for the first time. On the basis of these results, circ_0078607 expression may prove to be novel reliable prognosis factors for HGSOC patients.
Disclosure
The authors declare no conflicts of interest in this work.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (Grant No. 81572560 and Grant No. 82072866).
References
- Fidler MM, Bray F, Soerjomataram I. The global cancer burden and human development: a review. Scand J Public Health. 2018;46(1):27–36. doi:10.1177/1403494817715400
- Matulonis UA, Sood AK, Fallowfield L, Howitt BE, Sehouli J, Karlan BY. Ovarian cancer. Nat Rev Dis Primers. 2016;2:16061. doi:10.1038/nrdp.2016.6127558151
- Eisenhauer EA. Real-world evidence in the treatment of ovarian cancer. Ann Oncol. 2017;28(suppl_8):viii61–viii65. doi:10.1093/annonc/mdx44329232466
- Lu KH. Screening for ovarian cancer in asymptomatic women. JAMA. 2018;319(6):557–558. doi:10.1001/jama.2017.2189429450509
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature1192823446348
- Wu J, Jiang Z, Chen C, et al. CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Lett. 2018;430:179–192. doi:10.1016/j.canlet.2018.05.03329803789
- Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6. doi:10.1186/s12943-018-0934-630626395
- Lu Q, Liu T, Feng H, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18(1):111. doi:10.1186/s12943-019-1040-031228937
- Chen N, Zhao G, Yan X, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19(1):218. doi:10.1186/s13059-018-1594-y30537986
- Yin W, Xu J, Li C, Dai X, Wu T, Wen J. Circular RNA circ_0007142 facilitates colorectal cancer progression by modulating CDC25A expression via miR-122-5p. Onco Targets Ther. 2020;13:3689–3701. doi:10.2147/OTT.S23833832431519
- Ning L, Long B, Zhang W, et al. Circular RNA profiling reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers in epithelial ovarian cancer. Int J Oncol. 2018;53(6):2637–2646. doi:10.3892/ijo.2018.456630272264
- Yalan S, Yanfang L, He C, Yujie T. Circular RNA circRHOBTB3 inhibits ovarian cancer progression through PI3K/AKT signaling pathway. Panminerva Med. 2020. doi:10.23736/S0031-0808.20.03957-9
- Zhao Y, Qin XP, Lang YP, Kou D, Shao ZW. Circular RNA circ-SMAD7 promoted ovarian cancer cell proliferation and metastasis by suppressing KLF6. Eur Rev Med Pharmacol Sci. 2020;24(14):7563.32744668
- Liu N, Zhang J, Zhang LY, Wang L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(12):3713–3718. doi:10.26355/eurrev_201806_1525029949144
- Zou T, Wang PL, Gao Y, Liang WT. Circular RNA_LARP4 is lower expressed and serves as a potential biomarker of ovarian cancer prognosis. Eur Rev Med Pharmacol Sci. 2018;22(21):7178–7182. doi:10.26355/eurrev_201811_1625030468459
- Zhang N, Jin Y, Hu Q, et al. Circular RNA hsa_circ_0078607 suppresses ovarian cancer progression by regulating miR-518a-5p/Fas signaling pathway. J Ovarian Res. 2020;13(1):64. doi:10.1186/s13048-020-00664-132503653
- Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet. 2019;393(10177):1240–1253. doi:10.1016/S0140-6736(18)32552-230910306
- Au KK, Josahkian JA, Francis JA, Squire JA, Koti M. Current state of biomarkers in ovarian cancer prognosis. Future Oncol. 2015;11(23):3187–3195. doi:10.2217/fon.15.25126551891
- Xie J, Wang S, Li G, et al. circEPSTI1 regulates ovarian cancer progression via decoying miR-942. J Cell Mol Med. 2019;23(5):3597–3602. doi:10.1111/jcmm.1426030887698
- Zhang M, Xia B, Xu Y, Zhang Y, Xu J, Lou G. Circular RNA (hsa_circ_0051240) promotes cell proliferation, migration and invasion in ovarian cancer through miR-637/KLK4 axis. Artif Cells Nanomed Biotechnol. 2019;47(1):1224–1233. doi:10.1080/21691401.2019.159399930945557
- Chen H, Mao M, Jiang J, Zhu D, Li P. Circular RNA CDR1as acts as a sponge of miR-135b-5p to suppress ovarian cancer progression. Onco Targets Ther. 2019;12:3869–3879. doi:10.2147/OTT.S207938
- Chen Q, Zhang J, He Y, Wang Y. hsa_circ_0061140 Knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol Ther Nucleic Acids.2018;13:55–63. doi:10.1016/j.omtn.2018.08.010
- Chen Y, Ye X, Xia X, Lin X.Circular RNA ABCB10 correlates with advanced clinicopathological features and unfavorable survival, and promotes cell proliferation while reduces cell apoptosis in epithelial ovarian cancer. Cancer Biomark. 2019;26(2):151-161. doi:10.3233/CBM-190064
- Gao Y, Zhang C, Liu Y, Wang M. Circular RNA profiling reveals circRNA1656 as a novel biomarker in high grade serous ovarian cancer. Biosci Trends. 2019;13(2):204-211. doi:10.5582/bst.2019.01021
