Abstract
Purpose
Fibroblast growth factor receptor 3 (FGFR3) alterations are frequent in non-muscle-invasive bladder cancer (NMIBC), although current data regarding the prognostic and therapeutic relevance are inconsistent. We analyzed the prognostic role of FGFR3 mRNA expression in stage T1 NMIBC.
Patients and Methods
The mRNA expression of FGFR3 and cyclin-dependent kinase inhibitor 2A (CDKN2A) was measured by RT-qPCR in 80 patients with stage T1 NMIBC treated with transurethral resection of the bladder and correlated with clinical data and KRT5 and KRT20 expression, used as surrogate markers for basal and luminal subtypes, respectively.
Results
FGFR3 and CDKN2A transcript levels were not correlated. FGFR3 expression was associated with the expression of KRT5 (p=0.002) and KRT20 (p < 0.001). CDKN2A expression was negatively correlated with KRT5 (p=0.030). In Kaplan–Meier analysis and univariable Cox regression analysis, high FGFR3 expression was associated with significantly reduced recurrence-free survival (RFS) (HR=3.78; p < 0.001) and improved overall survival (OS) (HR=0.50; p=0.043), while high CDKN2A expression was associated with reduced OS (HR=2.34; p=0.034). Patient age was the only clinicopathological parameter associated with reduced OS (HR=2.29; p=0.022). No parameter was an independent prognostic factor in multivariable analysis. Next, we stratified the patients depending on their lineage differentiation. In univariable analysis, the prognostic effect of FGFR3 and CDKN2A was observed primarily in patients demonstrating high expression of KRT5 or KRT20, whereas high FGFR3 expression was associated with significantly reduced RFS, irrespective of instillation therapy.
Conclusion
Stage T1 NMIBC patients with high FGFR3 expression show shorter RFS but better OS than patients with low FGFR3 expression.
Introduction
Urothelial carcinoma of the bladder (UCB) is the 11th most common cancer worldwide.Citation1 Approximately 75% of newly diagnosed UCBs are non-muscle-invasive bladder cancers (NMIBC), which include stages Ta, T1 and carcinoma in situ (CIS). NMIBCs are usually treated with a bladder-sparing approach comprising transurethral resection of the bladder (TUR-B) followed by regular cystoscopies. As an adjuvant treatment, instillation therapies of mitomycin or Bacillus Calmette-Guérin (BCG) can be administered, depending on tumor status.Citation2 High rates of recurrence and progression to muscle-invasive bladder cancer (MIBC) necessitate frequent follow-up examinations, which pose a heavy burden for the patient as well as the health care system.Citation3,Citation4 Although there have been some improvements in the treatment of metastatic disease in recent years, with the introduction of numerous checkpoint inhibitors as well as the accelerated approval of erdafitinib by the Food and Drug Administration (FDA), no new diagnostic methods or therapeutic options have been established for NMIBC.Citation5,Citation6 The lack of innovation is especially critical for patients with stage T1 NMIBC, as 70% experience a disease recurrence after BCG, while 33% of these patients even progress to MIBC.Citation7 Patients classified as having the highest risk of NMIBC according to the criteria developed by the European Organization for Research and Treatment of Cancer (EORTC) should be considered for an early cystectomy.Citation8 The investigation and clinical implementation of novel molecular markers beyond the established clinicopathological characteristics could be a helpful addition to better distinguish patients with stage T1 NMIBC suitable either for a bladder-sparing approach or early cystectomy.
Aberrations such as mutations or overexpression of the fibroblast growth factor receptor 3 (FGFR3) are very frequent in UCB with mutations occurring in approximately 40% of patients, of whom 70–80% have low-grade NMIBC, which allows ligand-independent dimerization, phosphorylation and downstream signaling.Citation9 Given the high frequency of FGFR3 mutations in urothelial papilloma and hyperplasia, both of which are considered precursors of papillary UCB, FGFR3 mutations supposedly occur early in the process of tumor development.Citation10 An association between FGFR3 mutations and lower stage and grade has been shown in several studies,Citation11–Citation13 which is also the case for patients with FGFR3 overexpression.Citation14,Citation15 Moreover, FGFR3 overexpression was previously associated with increased FGFR3 mutation rates. However, approximately 40% of UCBs with FGFR3 overexpression do not harbor any FGFR3 mutations, many of which are MIBCs.Citation16
Nevertheless, the prognostic relevance of FGFR3 with regard to survival in NMIBC remains unclear. In terms of cancer recurrence, several studies have demonstrated an association with reduced recurrence-free survival (RFS),Citation17 while others have found FGFR3 mutations to be associated with lower recurrence rates.Citation11 Regarding the association with progression-free survival (PFS), the results are conflicting as well. For instance, Burger et al found FGFR3 mutations to be associated with longer PFS,Citation13 unlike Hernández et al, who found no prognostic relevance of FGFR3 mutations.Citation18 These conflicting results are especially apparent in stage T1 NMIBC, which may be due to the many molecular similarities this tumor entity shares with stage Ta NMIBC as well as MIBC.Citation9,Citation19 The combination of FGFR3 expression with other markers might be necessary to improve the predictive value of FGFR3 expression in stage T1 NMIBC.
To overcome the known confinements of immunohistochemistry, we recently investigated the mRNA expression of FGFR3 with reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) assessment in a large cohort of 296 patients with stage T1 NMIBC and found an association of high FGFR3 expression with improved PFS.Citation20 In addition to FGFR3, we also measured the mRNA expression of cyclin-dependent kinase inhibitor 2A (CDKN2A), which encodes the tumor suppressor protein p16.Citation21 In other studies, a loss of heterozygosity in the region 9p of chromosome 9, and thereby the deletion of CDKN2A, has been associated with a higher grade and worse outcome.Citation22,Citation23 Interactions between CDKN2A and FGFR3 have previously been suggested by other investigators.Citation24 In our previous study, high mRNA expression of CDKN2A was associated with reduced PFS, with the subgroup of patients with high CDKN2A and low FGFR3 expression displaying the worst PFS.Citation20
The goal of the current study was to validate the prognostic relevance of the mRNA expression of FGFR3 and CDKN2A within a new independent cohort of patients with stage T1 NMIBC in order to allow for a future implementation into the diagnostics and therapy in a daily clinical routine.
Patients and Methods
Patient Population
In total, the clinical and histopathological data of 80 patients treated with TUR-B at the Department of Urology and Pediatric Urology of the University Hospital Erlangen between 2000 and 2015 were retrospectively analyzed. Only patients initially diagnosed with stage T1 NMIBC and treated with a bladder-sparing approach were included in this study. All patients received a Re-TUR-B within six to eight weeks after the initial TUR-B. Tumor tissue slices of all patients were evaluated for pathological stage according to the 2010 TNM classification and graded according to the common grading systems (WHO 1973, WHO 2016) by two experienced uropathologists (ME, AH). All specimens contained at least 50% tumor cells. All patients gave informed consent. All procedures were performed in accordance with the ethical standards established in the 1975 Declaration of Helsinki. The study was approved by the Ethics Committee of the University Hospital Erlangen (No. 3755 and No. 296_18 Bc). Recurrence was defined as the reappearance of UCB, either NMIBC or MIBC, while progression was defined as the progression to MIBC or metastatic disease.
Assessment of mRNA Expression by RT-qPCR
Tumor specimens were assessed by RT-qPCR as previously described.Citation20 In short, after extraction from a single 10-μm curl of FFPE tissue, the RNA was then processed according to a commercially available bead-based extraction method (Xtract kit; STRATIFYER Molecular Pathology GmbH, Cologne, Germany). RNA was eluted with 100 μL of elution buffer. DNA was digested, and RNA eluates were then stored at −80°C until use.
The mRNA expression levels of FGFR3 and CDKN2A were assessed, in addition to the keratins KRT5 and KRT20 as surrogate markers for basal and luminal markers of UCB, similar to our previous studies.Citation25,Citation26 Furthermore, mRNA expression of the proliferation marker Ki-67 (MKI67) was measured. Calmodulin-2 (CALM2) and ß2-microglobulin (B2M) were used as reference genes. The mRNA expression was determined by a one-step reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR)-based assessment, which involves the reverse transcription of RNA and subsequent amplification of cDNA executed in a one-step reaction. Each patient sample or control was analyzed in duplicate using the Invitrogen SuperScript III RT-qPCR system (Thermo Fisher Scientific, Waltham, MA, USA) and gene-specific primer-probe combinations (STRATIFYER Molecular Pathology). Each patient’s sample was analyzed in duplicate on an ABI Step One PCR System (Thermo Fisher Scientific) according to the manufacturer’s instructions. Gene expression was quantified with a modification of the method by Schmittgen and Livak by calculating 40-ΔCt, whereas ΔCt was calculated as the difference in Ct between the test gene and the mean of the reference genes.Citation20
Statistical Methods
Correlations between the mRNA expressions of FGFR3, CDKN2A, KRT5, KRT20, and MKI67 were calculated using Spearman’s bivariate correlation. Optimized cut-off values for each marker with regard to survival were defined using Youden’s index on the receiver operating characteristic (ROC) curve. The date of the first TUR-B was defined as the common time point zero for retrospective survival analysis. The associations of clinicopathological markers (Grade WHO 1973, concomitant carcinoma in situ (CIS), instillation therapy, gender, age) and mRNA expression of the molecular markers with RFS, PFS, overall survival (OS), and cancer-specific survival (CSS) were determined by univariable (Kaplan–Meier analysis and Cox regression hazard models) and multivariable analyses (Cox regression hazard models). All tests were two-sided, and p-values <0.05 were considered statistically significant. Statistical analyses were performed with the SPSS 21.0 software package (SPSS Inc., Chicago, IL, USA) and R V3.2.1 (The R foundation for statistical computing, Vienna, Austria).
Results
Patient Population
The clinicopathological characteristics of the cohort are summarized in . Three quarters of patients were males. The median age at diagnosis was 71 years (46–97), and the median follow-up was 62 months (range 0–189 months).
Table 1 Patient Cohort (IQR=Interquartile Range)
Almost half of the patients (51.2%) received an adjuvant instillation therapy with either mitomycin or BCG, which is comparable to real-world data showing an application of postoperative instillation therapy in 29–65% of patients with high-risk NMIBC.Citation27,Citation28
Correlation of the mRNA Expression of FGFR3, CDKN2A, KRT5, KRT20, and MKI67 with Each Other and with Clinicopathological Parameters
Nonparametric Spearman’s rank test revealed no correlation between the mRNA expression of FGFR3 and CDKN2A. CDKN2A showed a significant negative correlation with KRT5 (rs= −0.24; p=0.030), ie, the basal subtype. In contrast, there was no correlation with KRT20, ie, the luminal subtype. FGFR3 was significantly associated with both KRT5 (rs=0.35; p=0.002) and KRT20 (rs=0.39; p < 0.001). Neither FGFR3 nor CDKN2A was significantly associated with MKI67. High FGFR3 expression was negatively correlated with tumor grade according to the WHO 1973 classification (rs= −0.33; p < 0.001) and concomitant CIS (rs= −0.33; p < 0.001). There was no association between FGFR3 or CDKN2A and age, gender, or instillation therapy.
Association of mRNA Expression of FGFR3, CDKN2A and Clinicopathological Parameters with Survival
We used receiver-operating characteristic (ROC) analyses to determine the optimal cut-off values for FGFR3 and CDKN2A with regard to survival (). For each clinical endpoint (OS, CSS, RFS, PFS) an optimal cut-off value was determined by the Youden index and applied in Kaplan Meier analyses (log rank test). Kaplan–Meier analysis showed high mRNA expression of FGFR3 to be significantly associated with reduced RFS (p < 0.001) and improved OS (p=0.039), in addition to showing a nonsignificant trend towards improved CSS (p=0.067). Interestingly, high FGFR3 expression was also associated with prolonged PFS (p=0.037); however, given that only six patients (7.5%) had a documented time of progression despite 16 patients suffering a cancer-specific death, we excluded PFS from any further survival analyses.
Table 2 Results of Kaplan–Meier Analysis (Log Rank Test): Cut-off Values for FGFR3 and CDKN2A with Regard to Recurrence-Free (RFS), Progression-Free (PFS), Cancer-Specific (CSS) and Overall Survival (OS) for the Total Cohort (n=80)
High expression of CDKN2A was significantly associated with reduced OS (p=0.029) and showed a nonsignificant trend towards reduced CSS (p=0.057). There was no association between mRNA expression of CDKN2A and RFS or PFS.
In the univariable Cox regression analysis, high FGFR3 expression was associated with a significantly increased risk of recurrence (hazard ratio (HR)=3.78; p < 0.001) and an improved chance for prolonged OS (HR=0.50; p=0.043). High CDKN2A was associated with a higher risk of reduced OS (HR=2.34; p=0.034).
Of the clinicopathological parameters, only age was associated with shorter CSS (HR=3.44; p=0.034) and OS (HR=2.29; p=0.022) in the univariable Cox regression analysis. Tumor grade; concomitant CIS; instillation therapy; gender; and the molecular parameters MKI67, KRT5, and KRT20 were not associated with prognosis (OS, CSS, RFS) and therefore were not included in further multivariable Cox regression analyses.
The multivariable analysis of OS adjusted for age and the expressions of FGFR3 and CDKN2A revealed that none of the parameters were independent prognostic factors. Given that FGFR3 expression was the only significant prognostic marker for RFS, no multivariable analysis was conducted for FGFR3.
Association of FGFR3 and CDKN2A mRNA Expression with RFS and OS Stratified by Clinicopathological Parameters or mRNA Expression
Having found an association between RFS and mRNA levels of FGFR3 as well as between RFS and both FGFR3 and CDKN2A in the total cohort, we sought to analyze the prognostic relevance of these two markers within different patient subgroups defined by clinicopathological parameters.
Stratification by Age
Using the median age of 71 years as a cut-off to define the two age groups (≤71 vs >71 years), patients aged ≤71 years who had NMIBC with high FGFR3 expression demonstrated significantly improved OS when compared to patients with low FGFR3 expression (p=0.007; Log rank test) (). In the univariable Cox regression analysis, patients aged ≤71 years with a high FGFR3 expression had a significantly reduced risk of death (HR=0.22; p=0.013) (). In patients older than 71 years, FGFR3 expression had no effect on OS. With regard to RFS, high FGFR3 expression was significantly associated with a shorter time to recurrence irrespective of the patients’ age (≤71 years: p=0.004; >71 years: p=0.010; Log rank test). The risk of experiencing a recurrence was also significantly increased with high FGFR3 expression in both age groups (≤71 years HR=4.87; p=0.008; >71 years HR=3.08; p=0.015) () in the univariable Cox regression analysis.
Table 3 Univariable Cox Regression Analysis for Stratification by Clinicopathological or Molecular Parameters: The Association of FGFR3 and CDKN2A mRNA with OS and RFS
Figure 1 Kaplan-Meier analysis of FGFR3 mRNA expression regarding RFS in patients ≤71 years (A) and patients >71 years (B) as well as regarding OS in patients ≤71 years (C) and patients >71 years (D).
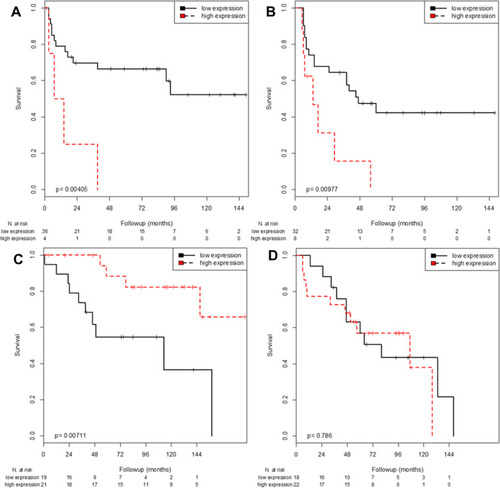
CDKN2A demonstrated no association with OS when the cohort was separated into the two age groups.
Stratification by KRT5 and KRT20 Expression
Both KRT5 and KRT20 mRNA expressions were used as surrogate markers to define basal and luminal subtypes of UCB by analogy to previous studies.Citation25 Median expression levels were used as cut-offs to subdivide patients into low/high KRT5 (≤36.78 vs >36.78) or low/high KRT20 (≤37.47 vs >37.47) mRNA expression.
As mentioned above, FGFR3 was associated with both markers. Patients with high expression of KRT5 were associated with reduced RFS (p < 0.001; Log rank test) and improved OS (p=0.023, Log rank test) when FGFR3 expression was high. In these patients, FGFR3 was also associated with an increased risk of recurrence (HR=4.92; p < 0.001) () but a better chance of improved OS (HR=0.27; p=0.032) in the univariable Cox regression analysis. In patients with low KRT5 expression, FGFR3 expression showed no effect on survival. In patients with high KRT20 expression, high FGFR3 expression was associated with reduced RFS (p=0.004; Log rank test; HR=3.43, p=0.007; univariable Cox regression analysis) () but not OS.
While CDKN2A showed no positive association with either KRT5 or KRT20 in the total cohort, patients with high expression of both markers showed a significantly reduced OS when CDKN2A expression was high (KRT5 p < 0.001; KRT20 p=0.039) (). CDKN2A was an independent prognostic marker for reduced OS in patients with high expression of KRT5 (HR=9.06; p=0.005) and KRT20 (HR=3.03; p=0.049) (). No effect of CDKN2A on OS was observed when the mRNA expression of KRT5 and KRT20 was low.
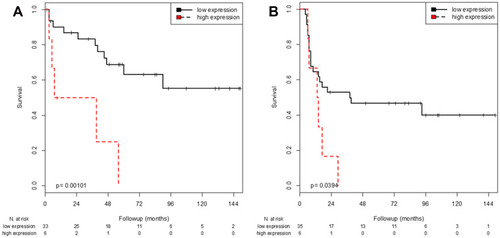
Figure 2 Kaplan-Meier analysis of FGFR3 mRNA expression regarding RFS in patients who received no postoperative instillation (A) and patients with postoperative instillation (B).
Combined, the current results indicate a prognostic effect of high FGFR3 or CDKN2A expression, especially in patients also presenting with either a basal or luminal-like subtype, while no prognostic relevance was seen in patients who could not be assigned to either of these subtypes.
Stratification by MKI67 Expression
MKI67 is a prominent marker associated with the proliferation activity of tumor cells.Citation29 As with KRT5 and KRT20, patients were divided into groups with low or high mRNA expression of MKI67 using the median expression level (≤33.10 vs >33.10). This way, patients with both low and high MKI67 expression were associated with significantly reduced RFS (MKI67 low p=0.013; MKI67 high p=0.010; Log rank test) when FGFR3 expression was high, and there was an increased risk of recurrence in both groups (MKI67 low: HR=4.59; p=0.022; MKI67 high: HR=2.99; p=0.014; univariable Cox regression analysis) (), unlike patients with low expression of FGFR3. There was no association with OS irrespective of MKI67 expression or the expression of either FGFR3 or CDKN2A. These data suggest that MKI67 expression plays no relevant role in the increased risk of recurrence for patients with high FGFR3 expression.
Stratification by Instillation
Independent of intravesical instillation therapy, patients with high expression of FGFR3 showed a shorter RFS (no instillation: p=0.001; installation: p=0.039; Log rank test) () and had an increased risk of recurrence. Nevertheless, the risk was higher for patients not receiving an instillation therapy (no instillation: HR=5.44, p=0.003; instillation: HR=2.61, p=0.047; univariable Cox regression analysis) (). There was no effect of instillation therapy on OS irrespective of FGFR3 or CDKN2A expression.
Discussion
Aberrations of FGFR3 are regarded as changes in one of the major pathways in the carcinogenesis of UCB.Citation30 Consequently, the role and effect of FGFR3 in UCB have been the focus of multiple studies.Citation12,Citation13,Citation21,Citation31 Most studies have found that FGFR3 alterations, such as mutations and protein overexpression, are associated with lower stage and grade and improved outcome, although the association with survival has not been consistent.Citation11,Citation13,Citation30,Citation32 For instance, initial analyses by van Rhijn et al demonstrated FGFR3 mutations to be associated with prolonged RFS and CSS.Citation17,Citation30 By contrast, Hernández et al found FGFR3 mutations to be associated with increased recurrence rates, but only in stage TaG1 NMIBC.Citation11 Other studies found no association with recurrence, progression or CSS.Citation11,Citation33,Citation34 Burger et al analyzed 221 patients with NMIBC and showed that FGFR3 mutations were associated with favorable PFS, especially in high-grade and stage T1 NMIBC.Citation13 This is in line with our previous results showing that FGFR3 mRNA overexpression is associated with improved PFS in 296 patients with stage T1 NMIBC as well as with our current results.Citation20 The improved outcome is also reflected in the improved OS demonstrated in our current study; however, this is only relevant in younger patients, suggesting that factors other than FGFR3 expression are more substantial in elderly patients. Higher patient age has generally been associated with reduced RFS in a cohort with primary CIS.Citation35 However, in our cohort of stage pT1 patients, we were not able to detect such an association. Instead, only high FGFR expression was accompanied by reduced RFS, implicating recurring but not very aggressive cancers irrespective of patients’ age. Recently, a meta-analysis showed that both FGFR3 mutation and protein overexpression were significantly associated with RFS, PFS, CSS, and overall survival in NMIBC.Citation36
While the overall benign effect of FGFR3 mutations in UCB is widely assumed, several studies have examined additional alterations or mutations to improve the stratification of patients with an increased risk of progression. Homozygous deletions of 9p21.3, including the CDKN2A gene, have been detected by array-based comparative genomic hybridization and/or RT-qPCR in 22% (9/41) of bladder cancer patients.Citation37 Several groups have investigated FGFR3 mutation in combination with a loss of heterozygosity in the 9p region of chromosome 9, which leads to a deletion of CDKN2A and decreased expression of the tumor suppressor protein p16, consequently promoting tumor progression.Citation38,Citation39 Ploussard et al investigated 58 patients with NMIBC for their CDKN2A and FGFR3 status.Citation24 In patients who retained heterozygosity on chromosome 9, FGFR mutational status was not predictive of recurrence or progression to MIBC. On the other hand, FGFR3 mutational status was strongly associated with outcomes in patients with a loss of heterozygosity, with patients with wild-type FGFR3 exhibiting a higher risk for recurrence and progression than patients with FGFR3 mutations.Citation24 Rebouissou et al found homozygous deletions of CDKN2A to be more frequent in patients with FGFR3-mutated UCB.Citation39 By analyzing 19 patients with NMIBC and grade heterogeneity, Downes et al found that NMIBCs with FGFR3 mutations demonstrated homozygous deletions of CDKN2A in particular in the low-grade regions of the tumor, suggesting that a loss of CDKN2A precedes grade progression.Citation40
Drawing conclusions from these studies, a worse outcome might be expected in patients with reduced CDKN2A expression. Interestingly, our previous study showed high mRNA expression of CDKN2A to be associated with reduced PFS, with the subgroup of patients with high CDKN2A and low FGFR3 expression displaying the worst PFS.Citation20 The negative effect of high CDKN2A expression in stage T1 NMIBC is in line with our current results, where we found high CDKN2A to be significantly associated with reduced OS and strongly trending towards reduced CSS. In a previous analysis of CDKN2A mRNA expression in patients with MIBC, high expression was associated with reduced RFS and CSS.Citation41 Moreover, in MIBC, very high or very low CDKN2A expression can be a predictor of worse survival.Citation42 Previous gene expression analyses displayed distinct mRNA expression patterns for stage Ta NMIBC and MIBC, with stage T1 NMIBC showing signatures of either one or the other.Citation43 Concerning CDKN2A expression, stage T1 NMIBC might resemble stage Ta NMIBC more than MIBC.
However, it is still unclear why increased CDKN2A mRNA expression appears to be mostly associated with a poor prognosis, whereas CDKN2A is considered to be a tumor suppressor gene.Citation44 Point mutations in CDKN2A do not play a role in UCB; however, CDKN2A functions upstream of the tumor suppressor RB1, and its expression is related to TP53 expression.Citation45,Citation46 Mutations in TP53 or RB1 can attenuate the effect of CDKN2A. In The Cancer Genome Atlas (TCGA) data TP53 mutations were observed in 49% and RB1 mutations in 13% of NMIBC patients, and even higher rates have been described by Meeks et al.Citation47,Citation48 However, this is controversial, since Heedegard et al identified only 8% and 7% mutations in the TP53 and RB1 genes, respectively, in their NMIBC cohort.Citation49 Furthermore, the relationship between CDKN2A and FGFR3 remains unclear. Al-Khalaf et al showed that CDKN2A can upregulate the expression of several genes involved in cell proliferation, such as fibroblast growth factor receptor 1 (FGFR1), cyclin D1 (CCND1) and E2F1 transcription factor 1 (E2F1). In this way, E2F1 mediates the p16-dependent regulation of several pro- and anti-apoptotic proteins.Citation50 However, a direct relationship has not yet been shown between CDKN2A and FGFR3. Interestingly, CDKN2A can positively regulate the senescence-associated microRNAs miR-26b, −181a, −210 and −424.Citation51 An in silico prediction program (TargetScan: http://www.targetscan.org/cgi-bin/targetscan/vert_72/) predicts that miR-181-5p and miR-424-5p can negatively regulate FGFR3. However, further research is necessary to study the possible relationship between CDKN2A and FGFR3.
Regarding the association of FGFR3 with molecular subtypes, the currently available data do not allow for final conclusions. Sjödahl et al showed a high FGFR3 protein expression in the urobasal A subtype,Citation52 which was in concordance with our previous results showing a correlation of FGFR3 mRNA expression with KRT5.Citation20 Hurst and Knowles found FGFR3 alterations mainly in the luminal-papillary subtype of MIBC, which is associated with the best overall survival.Citation53 In the present study, FGFR3 mRNA expression was significantly associated with both KRT5 and KRT20, suggesting that high FGFR3 expression cannot be generally assigned to either the basal or luminal subtype. This may also be due to the special nature of T1 NMIBC, as it can exhibit molecular signatures of either NMIBC or MIBC.Citation9,Citation43 Intriguingly, when stratifying patients by their KRT5 and KRT20 expression, a prognostic effect of FGFR3 and CDKN2A was observed only when one of the keratins was expressed at high levels. This is indicative of an association of FGFR3 and CDKN2A with either a basal or luminal-like subtype, unlike other possible subtypes that do not express KRT5 or KRT20.
In several studies reporting on the response to FGFR inhibitors (in MIBC), complete response rates, disease control rates, and overall response rate of 0% to 8%, 59.3% to 64.2%, and 40% were reported for dovitinib, infigratinib, and erdafitinib, respectively.Citation36 However, the therapeutic consequences of FGFR3 inhibitors in NMIBC remain disputed. Although FGFR3 mutations are mainly associated with NMIBC, no targeted therapies have been approved yet. Our current results demonstrate reduced RFS but prolonged OS in stage T1 NMIBC with high FGFR3 expression, indicating less aggressive but frequently recurring tumors that might benefit from therapies that reduce recurrence rates. Interestingly, while patients with high FGFR3 expression had significantly shorter RFS than patients with low FGFR3 expression, regardless of instillation therapies, the risk of recurrence was five-fold higher when no instillation was applied compared to only approximately two-fold with instillation therapy. These data suggest that patients with high FGFR3 expression still might benefit from a conventional intravesical instillation therapy, which might be improved in combination with targeted therapies. Thus far, however, the sole implementation of anti-FGFR3 therapies in NMIBC has not been successful. In a recent Phase II trial, patients with NMIBC unresponsive to BCG received oral dovitinib.Citation54 While pharmacodynamically active dovitinib concentrations were observed in urothelial tissues in all patients, over 90% of patients showed no response to therapy, with all patients experiencing at least one grade 3 or 4 toxicity. Currently, ClinicalTrials.gov lists two phase II clinical trials that are still recruiting patients. One study is investigating the antineoplastic effect of the FGFR inhibitor pemigatinib as well as the therapeutic relevance of FGFR3 alterations in patients with recurrent low or intermediate risk NMIBC prior to second TUR-B (NCT03914794). The other study is examining the effect of erdafitinib versus either gemcitabine or mitomycin in patients with high-risk NMIBC and FGFR3 alterations with recurrence after BCG (NCT04172675).
The limitations of the current study include the retrospective nature as well as the relatively small cohort of 80 patients, which limits the reproducibility. In addition, no further subclassification with regard to the combined FGFR3 and CDKN2A expression was performed, as the small subgroups would not allow any meaningful conclusions. No immunohistochemistry was used, which is the most common method of marker quantification. There are several other biomarkers described with prognostic and/or predictive impact for NMIBC that were not included in this study.Citation36,Citation55 Moreover, only mRNA expression levels and no mutational status was analyzed. However, a recent study in stage T1 NMIBC showed an association between FGFR3 mutations and higher expression of FGFR3 mRNA.Citation56 Finally, while we validated the association of clinicopathological features with FGFR3 and CDKN2A mRNA expression. However, we did not aim to replicate individual thresholds, although the interlab variation for mRNA analysis appeared to be reasonably low.Citation57 We suggest that future prospective clinical trials may determine valid thresholds for better implementation of mRNA quantification into daily clinical practice. Altogether, we could verify the association of FGFR3 and CDKN2A mRNA expression with long-term prognostic outcomes in NMIBC patients.
Conclusion
In conclusion, we were able to confirm the overall positive prognostic role of high FGFR3 mRNA expression in stage T1 NMIBC, although these tumors are associated with increased recurrence rates. A conclusive assignment to either basal or luminal subtypes is not possible in stage T1 NMIBC. High CDKN2A expression is associated with unfavorable outcomes. Additional studies are necessary to investigate the applicability and usefulness of anti-FGFR3-targeted therapies in the difficult-to-treat stage T1 NMIBC.
Disclosure
DS, HT, JB, ME, VW, BK, WO, TSW, MCK, PE, AH, BW, RMW and SW are members of the BRIDGE Consortium e.V., 68167 Mannheim, Germany. ME reports grants, personal fees, and/or non-financial support from AstraZeneca, Janssen, Cepheid, MSD, Roche, Astellas, GenomicHealth, Diaceutics, and STRATIFYER, outside the submitted work. JK reports grants from ELAN Fund, during the conduct of the study. AH reports personal fees from BMS, MSD, Roche, Janssen, Pfizer, AstraZeneca, Cepheid, and Qiagen, during the conduct of the study; personal fees from BMS, MSD, Roche, AstraZeneca, Janssen, Qiagen, and Cepheid, outside the submitted work. RMW reports fee for service research cooperation from Janssen Research & Development LLC, fee for master research and testing service collaboration from Qiagen GmbH, during the conduct of the study; is employee and reports stocks from STRATIFYER Molecular Pathology, fee for strategic framework collaboration from BioNTech Diagnostics GmbH, outside the submitted work; In addition, RMW has a patent “Method of classifying a sample based on determination of FGFR” (PCT/EP2020/060456) licensed to Qiagen GmbH. The authors report no other conflicts of interest in this work.
Acknowledgments
The authors thank Angela Neumann for excellent technical support. The authors thank American Journal Experts for editing the manuscript. The authors also acknowledge support by the Deutsche Forschungsgemeinschaft and Friedrich-Alexander University Erlangen-Nuremberg, Erlangen, Germany within the funding program Open Access Publishing.
References
- FerlayJ, SoerjomataramI, DikshitR, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–386. doi:10.1002/ijc.2921025220842
- BabjukM, BurgerM, CompératEM, et al. European Association of urology guidelines on non-muscle-invasive bladder cancer (TaT1 and carcinoma in situ) - 2019 update. Eur Urol. 2019;76(5):639–657. doi:10.1016/j.eururo.2019.08.01631443960
- ProutGRJr, BartonBA, GriffinPP, FriedellGH; The National Bladder Cancer Group. Treated history of noninvasive grade 1 transitional cell carcinoma. J Urol. 1992;148(5):1413–1419. doi:10.1016/S0022-5347(17)36924-01433540
- HongYM, LoughlinKR. Economic impact of tumor markers in bladder cancer surveillance. Urology. 2008;71(1):131–135. doi:10.1016/j.urology.2007.08.01418242381
- WołącewiczM, HrynkiewiczR, GrywalskaE. Immunotherapy in bladder cancer: current methods and future perspectives. Cancers. 2020;12(5):1181. doi:10.3390/cancers12051181
- LoriotY, NecchiA, ParkSH, et al. Erdafitinib in locally advanced or metastatic urothelial carcinoma. N Engl J Med. 2019;381(4):338–348. doi:10.1056/NEJMoa181732331340094
- ShahinO, ThalmannGN, RentschC, MazzucchelliL, StuderUE. A retrospective analysis of 153 patients treated with or without intravesical bacillus Calmette-Guerin for primary stage T1 grade 3 bladder cancer: recurrence, progression and survival. J Urol. 2003;169(1):96–100; discussion 100. doi:10.1016/S0022-5347(05)64044-X12478112
- BabjukM, BohleA, BurgerM, et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: update 2016. Eur Urol. 2017;71(3):447–461. doi:10.1016/j.eururo.2016.05.041
- KnowlesMA. Novel therapeutic targets in bladder cancer: mutation and expression of FGF receptors. Future Oncol. 2008;4(1):71–83. doi:10.2217/14796694.4.1.7118241002
- KnowlesMA. FGFR3 – a central player in bladder cancer pathogenesis?Bladder Cancer. 2020;Preprint:1–21.
- HernándezS, López-KnowlesE, LloretaJ, et al. Prospective study of FGFR3 mutations as a prognostic factor in nonmuscle invasive urothelial bladder carcinomas. J Clin Oncol. 2006;24(22):3664–3671. doi:10.1200/JCO.2005.05.177116877735
- van RhijnBW, ZuiverloonTC, VisAN, et al. Molecular grade (FGFR3/MIB-1) and EORTC risk scores are predictive in primary non-muscle-invasive bladder cancer. Eur Urol. 2010;58(3):433–441. doi:10.1016/j.eururo.2010.05.04320646825
- BurgerM, van der AaMN, van OersJM, et al. Prediction of progression of non-muscle-invasive bladder cancer by WHO 1973 and 2004 grading and by FGFR3 mutation status: a Prospective Study. Eur Urol. 2008;54(4):835–843. doi:10.1016/j.eururo.2007.12.02618166262
- Gómez-RománJJ, SaenzP, MolinaM, et al. Fibroblast growth factor receptor 3 is overexpressed in urinary tract carcinomas and modulates the neoplastic cell growth. Clin Cancer Res. 2005;11(2 Pt 1):459–465.15701828
- Mhawech-FaucegliaP, CheneyRT, FischerG, BeckA, HerrmannFR. FGFR3 and p53 protein expressions in patients with pTa and pT1 urothelial bladder cancer. Eur J Surg Oncol. 2006;32(2):231–237. doi:10.1016/j.ejso.2005.11.01816412606
- TomlinsonDC, BaldoO, HarndenP, KnowlesMA. FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. J Pathol. 2007;213(1):91–98. doi:10.1002/path.220717668422
- van RhijnBW, LurkinI, RadvanyiF, KirkelsWJ, van der KwastTH, ZwarthoffEC. The fibroblast growth factor receptor 3 (FGFR3) mutation is a strong indicator of superficial bladder cancer with low recurrence rate. Cancer Res. 2001;61(4):1265–1268.11245416
- HernándezS, López-KnowlesE, LloretaJ, et al. FGFR3 and Tp53 mutations in T1G3 transitional bladder carcinomas: independent distribution and lack of association with prognosis. Clin Cancer Res. 2005;11(15):5444–5450. doi:10.1158/1078-0432.CCR-05-012216061860
- RichterJ, JiangF, GörögJP, et al. Marked genetic differences between stage pTa and stage pT1 papillary bladder cancer detected by comparative genomic hybridization. Cancer Res. 1997;57(14):2860–2864.9230190
- BreyerJ, WirtzRM, ErbenP, et al. High CDKN2A/p16 and low FGFR3 expression predict progressive potential of stage pT1 urothelial bladder carcinoma. Clin Genitourin Cancer. 2018;16(4):248–256.e242. doi:10.1016/j.clgc.2018.01.00929525349
- PollardC, SmithSC, TheodorescuD. Molecular genesis of non-muscle-invasive urothelial carcinoma (NMIUC). Expert Rev Mol Med. 2010;12:e10. doi:10.1017/S146239941000140720334706
- BerggrenP, KumarR, SakanoS, et al. Detecting homozygous deletions in the CDKN2A(p16(INK4a))/ARF(p14(ARF)) gene in urinary bladder cancer using real-time quantitative PCR. Clin Cancer Res. 2003;9(1):235–242.12538475
- AbatD, DemirhanO, InandikliogluN, et al. Genetic alterations of chromosomes, p53 and p16 genes in low- and high-grade bladder cancer. Oncol Lett. 2014;8(1):25–32. doi:10.3892/ol.2014.210824959214
- PloussardG, SolimanH, DubosqF, et al. The prognostic value of FGFR3 mutational status for disease recurrence and progression depends on allelic losses at 9p22. Am J Cancer Res. 2011;1(4):498–507.21984968
- BreyerJ, WirtzRM, OttoW, et al. In stage pT1 non-muscle-invasive bladder cancer (NMIBC), high KRT20 and low KRT5 mRNA expression identify the luminal subtype and predict recurrence and survival. Virchows Arch. 2017;470(3):267–274. doi:10.1007/s00428-017-2064-828074276
- EcksteinM, WirtzRM, Gross-WeegeM, et al. mRNA-expression of KRT5 and KRT20 defines distinct prognostic subgroups of muscle-invasive urothelial bladder cancer correlating with histological variants. Int J Mol Sci. 2018;19(11):11. doi:10.3390/ijms19113396
- van RhijnBW, BurgerM. Bladder cancer: low adherence to guidelines in non-muscle-invasive disease. Nat Rev Urol. 2016;13(10):570–571. doi:10.1038/nrurol.2016.16527578042
- IedaT, MutoS, ShimizuF, et al. Development and validation of a novel recurrence risk stratification for initial non-muscle invasive bladder cancer in Asia. EBioMedicine. 2016;12:98–104. doi:10.1016/j.ebiom.2016.08.05127614395
- GerdesJ, SchwabU, LemkeH, SteinH. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer. 1983;31(1):13–20. doi:10.1002/ijc.29103101046339421
- van RhijnBW, van der KwastTH, VisAN, et al. FGFR3 and P53 characterize alternative genetic pathways in the pathogenesis of urothelial cell carcinoma. Cancer Res. 2004;64(6):1911–1914. doi:10.1158/0008-5472.CAN-03-242115026322
- van OersJM, ZwarthoffEC, RehmanI, et al. FGFR3 mutations indicate better survival in invasive upper urinary tract and bladder tumours. Eur Urol. 2009;55(3):650–657. doi:10.1016/j.eururo.2008.06.01318584939
- Di MartinoE, TomlinsonDC, KnowlesMA. A decade of FGF receptor research in bladder cancer: past, present, and future challenges. Adv Urol. 2012;2012(429213):1–10. doi:10.1155/2012/429213
- HosenI, RachakondaPS, HeidenreichB, et al. Mutations in TERT promoter and FGFR3 and telomere length in bladder cancer. Int J Cancer. 2015;137(7):1621–1629. doi:10.1002/ijc.2952625809917
- ZiegerK, DyrskjøtL, WiufC, et al. Role of activating fibroblast growth factor receptor 3 mutations in the development of bladder tumors. Clin Cancer Res. 2005;11(21):7709–7719. doi:10.1158/1078-0432.CCR-05-113016278391
- KimSJ, NamW, YouD, et al. Prognostic factors related to recurrence-free survival for primary carcinoma in situ of the bladder after bacillus calmette-guérin: a Retrospective Study. Urol Int. 2018;101(3):269–276. doi:10.1159/00049212130179882
- Kardoust PariziM, MargulisV, LotanY, MoriK, ShariatSF. Fibroblast growth factor receptor: a systematic review and meta-analysis of prognostic value and therapeutic options in patients with urothelial bladder carcinoma. Urol Oncol. 2021;39(7):409–421. doi:10.1016/j.urolonc.2021.01.02533642228
- VeltmanJA, FridlyandJ, PejavarS, et al. Array-based comparative genomic hybridization for genome-wide screening of DNA copy number in bladder tumors. Cancer Res. 2003;63(11):2872–2880.12782593
- BartolettiR, CaiT, NesiG, Roberta GirardiL, BaroniG, Dal CantoM. Loss of P16 expression and chromosome 9p21 LOH in predicting outcome of patients affected by superficial bladder cancer. J Surg Res. 2007;143(2):422–427. doi:10.1016/j.jss.2007.01.01217612565
- RebouissouS, HéraultA, LetouzéE, et al. CDKN2A homozygous deletion is associated with muscle invasion in FGFR3-mutated urothelial bladder carcinoma. J Pathol. 2012;227(3):315–324. doi:10.1002/path.401722422578
- DownesMR, WeeningB, van RhijnBW, HaveCL, TreurnietKM, van der KwastTH. Analysis of papillary urothelial carcinomas of the bladder with grade heterogeneity: supportive evidence for an early role of CDKN2A deletions in the FGFR3 pathway. Histopathology. 2017;70(2):281–289. doi:10.1111/his.1306327530957
- WorstTS, WeisCA, StöhrR, et al. CDKN2A as transcriptomic marker for muscle-invasive bladder cancer risk stratification and therapy decision-making. Sci Rep. 2018;8(1):14383.30258198
- ShariatSF, TokunagaH, ZhouJ, et al. p53, p21, pRB, and p16 expression predict clinical outcome in cystectomy with bladder cancer. J Clin Oncol. 2004;22(6):1014–1024. doi:10.1200/JCO.2004.03.11814981102
- LindgrenD, FrigyesiA, GudjonssonS, et al. Combined gene expression and genomic profiling define two intrinsic molecular subtypes of urothelial carcinoma and gene signatures for molecular grading and outcome. Cancer Res. 2010;70(9):3463–3472. doi:10.1158/0008-5472.CAN-09-421320406976
- VogelsteinB, PapadopoulosN, VelculescuVE, ZhouS, DiazLAJr, KinzlerKW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi:10.1126/science.123512223539594
- LukasJ, ParryD, AagaardL, et al. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature. 1995;375(6531):503–506. doi:10.1038/375503a07777060
- PomerantzJ, Schreiber-AgusN, LiégeoisNJ, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92(6):713–723. doi:10.1016/S0092-8674(00)81400-29529248
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–322. doi:10.1038/nature1296524476821
- MeeksJJ, CarneiroBA, PaiSG, et al. Genomic characterization of high-risk non-muscle invasive bladder cancer. Oncotarget. 2016;7(46):75176–75184. doi:10.18632/oncotarget.1266127750214
- HedegaardJ, LamyP, NordentoftI, et al. Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell. 2016;30(1):27–42. doi:10.1016/j.ccell.2016.05.00427321955
- Al-KhalafHH, ColakD, Al-SaifM, et al. p16(INK4a) positively regulates cyclin D1 and E2F1 through negative control of AUF1. PLoS One. 2011;6(7):e21111. doi:10.1371/journal.pone.002111121799732
- OverhoffMG, GarbeJC, KohJ, StampferMR, BeachDH, BishopCL. Cellular senescence mediated by p16INK4A-coupled miRNA pathways. Nucleic Acids Res. 2014;42(3):1606–1618. doi:10.1093/nar/gkt109624217920
- SjödahlG, LaussM, LövgrenK, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res. 2012;18(12):3377–3386. doi:10.1158/1078-0432.CCR-12-0077-T22553347
- HurstCD, KnowlesMA. Bladder cancer: multi-omic profiling refines the molecular view. Nat Rev Clin Oncol. 2018;15(4):203–204. doi:10.1038/nrclinonc.2017.19529255240
- HahnNM, BivalacquaTJ, RossAE, et al. A phase II trial of dovitinib in BCG-unresponsive urothelial carcinoma with FGFR3 mutations or overexpression: Hoosier cancer research network trial HCRN 12-157. Clin Cancer Res. 2017;23(12):3003–3011. doi:10.1158/1078-0432.CCR-16-226727932416
- AfferiL, MoschiniM, CumberbatchMG, et al.; European Association of Urology - European Society of Resident Urologists (EAU-ESRU). Biomarkers predicting oncological outcomes of high-risk non-muscle-invasive bladder cancer. Minerva Urol Nefrol. 2020;72(3):265–278. doi:10.23736/S0393-2249.20.03786-832298067
- KangHW, KimYH, JeongP, et al. Expression levels of FGFR3 as a prognostic marker for the progression of primary pT1 bladder cancer and its association with mutation status. Oncol Lett. 2017;14(3):3817–3824. doi:10.3892/ol.2017.662128927152
- EcksteinM, WirtzRM, PfannstilC, et al. A multicenter round robin test of PD-L1 expression assessment in urothelial bladder cancer by immunohistochemistry and RT-qPCR with emphasis on prognosis prediction after radical cystectomy. Oncotarget. 2018;9(19):15001–15014. doi:10.18632/oncotarget.2453129599921
