Abstract
Purpose
Hsa_circ_0007637 was discovered to be differentially expressed in nasopharyngeal carcinoma (NPC). However, the exact function and mechanism of Hsa_circ_0007637 on NPC have not been studied. This study firstly researched the function and mechanism of Hsa_circ_0007637 on NPC progression.
Methods
Hsa_circ_0007637, miR-636 and TPD52 expressions in 80 NPC patients were detected by quantitative real-time polymerase chain reaction. Hsa_circ_0007637 effect on NPC cell proliferation, apopticosis, invasion and migration was investigated by cell counting kit-8 assay, flow cytometry, transwell experiment and wound healing assay accordingly. Dual-luciferase reporter gene assay, RNA immunoprecipitation experiment and RNA fluorescence in situ hybridization experiment were performed to identify the binding between Hsa_circ_0007637 and miR-636. Dual-luciferase reporter gene assay and RNA pull down assay were conducted to verify the binding between miR-636 and TPD52. TPD52 protein expression in NPC cells was determined by Western blot. In vivo study was performed using nude mice. Immunohistochemistry was performed to assess TPD52 and Ki67 expression in tissues.
Results
Hsa_circ_0007637 was overexpressed in NPC tissues and cells. High Hsa_circ_0007637 expression predicted a poor outcome for NPC patients. Hsa_circ_0007637 knockdown decreased proliferation, invasion, migration and increased apoptosis of NPC cells (P < 0.01). Hsa_circ_0007637 could enhance TPD52 expression via sponging miR-636. miR-636 overexpression or TPD52 knockdown weakened the promoting effect of Hsa_circ_0007637 on NPC cells malignant phenotype (P < 0.01). Hsa_circ_0007637 knockdown suppressed NPC cells growth in vivo (P < 0.01).
Conclusion
Hsa_circ_0007637 facilitates NPC progression by sponging miR-636/TPD52 axis.
Introduction
Nasopharyngeal carcinoma (NPC) is derived from the superior mucosal epithelium of the nasopharynx.Citation1 The incidence of NPC is affected by many factors, such as genetic predisposition, Epstein-Barr virus (EBV) infection and environmental factors.Citation2 The geographic distribution of NPC is very unique. Over 70% of new cases are in East and Southeast Asia.Citation3 The prevalence of NPC in Southern China is up to approximately 30 cases per 100,000 persons.Citation2 Great improvements have been achieved in the NPC treatment, but the 5-year survival rate of NPC patients still remains at about 70%. Even worse, many NPC patients suffer from recurrence or distant metastasis after treatment.Citation4 Hence, in order to improve the prognosis, the discovery of effective treatment targets for NPC is of great significance.
Circular RNAs (circRNAs) are a class of endogenous non-coding RNAs stably expressed in various organisms, which are characterized by covalently closed-loop structure.Citation1 Accumulated evidence shows that the abnormal expression of circRNAs is involved in the occurrence and development of multiple human malignant tumors through interfering with tumor cells proliferation, apoptosis, migration, invasion, cell cycle distribution, etc.Citation5,Citation6 A large number of studies have shown that circRNAs act as a competitive endogenous RNA (ceRNA) to sponge miRNAs and then regulate the expression of downstream target genes of miRNAs.Citation7,Citation8 The discovery of this mechanism provides a reliable theoretical basis for circRNAs as targets in tumor treatment. Although the function of several circRNAs in human tumors has been identified, a large amount of circRNAs function has not been fully explained currently. Recently, through high-throughput RNA sequencing, Wu et alCitation9 discovered that Hsa_circ_0007637 was differentially expressed in NPC tissues and cells. They speculated that Hsa_circ_0007637 might play a role in the development of NPC. However, the exact function of Hsa_circ_0007637 in NPC has not been elucidated. Thus, this study investigated the exact function of Hsa_circ_0007637 in NPC development. Through bioinformatics prediction, miR-636 was predicted to be a target gene of Hsa_circ_0007637 and tumor protein D52 (TPD52) was predicted to be a target gene of miR-636. Therefore, it was speculated that Hsa_circ_0007637 might regulate NPC development via targeting the miR-636/TPD52 axis. Detailed experiments were performed to verify the above hypothesis. The findings of this study will provide a promising target for NPC treatment.
Methods
Patients and Clinical Samples Collection
In this study, a total of 80 NPC patients voluntarily participated in this study. This study has been approved by the ethics committee of The First Affiliated Hospital of Soochow University. All patients have signed written informed consent. This study was conducted in accordance with the Declaration of Helsinki.
These patients were diagnosed with NPC for the first time. They experienced surgical resection in our hospital from 2012.3 to 2015.5. None of the patients have a previous treatment history of cancer-related diseases. The tumor tissues and matched adjacent normal tissues collected during surgery were immediately stored at −80°C. The clinical pathology of each patient was recorded, including ages, gender, distant metastasis, TNM Clinical stage and pathologic types. After treatment, all patients were followed up for 60 months.
Cell Lines and Culture
Normal nasopharyngeal epithelial cell line (NP460) and two NPC cell lines (C666-1 and 5–8F) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Dulbecco’s modified eagle medium (DMEM) containing 10% fetal bovine serum (FBS, Solarbio, Beijing, China) and antibiotics (100 U/mL penicillin and 100 mg/mL streptomycin, Solarbio, Beijing, China) was used to culture cells at 37°C, 5% CO2.
Ribonuclease R (RNase R) Treatment
Total RNAs were extracted by incubation 5–8F cells with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). A total of 5 μg RNA sample were collected and incubated with RNase R (3 U/μg, Epicentre, Madison, WI, USA) for 20 min at 37°C. The expression of Hsa_circ_0007637 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was then detected by quantitative reverse transcription polymerase chain reaction (qRT-PCR). Simultaneously, Hsa_circ_0007637 and GAPDH expressions in 5 μg RNA sample (without RNase R digestion) were used as the Mock. GAPDH was used as the representative linear RNA. The linear structure could be destroyed by RNase R, while the circle structure could not be destroyed by RNase R.
Cell Transfection
C666-1 and 5–8F cells were seeded in 6-well plates (1 × 106 cells per well) with DMEM (without FBS) for 8 h transfection at 37°C, 5% CO2. Briefly, Hsa_circ_0007637 siRNA (siRNA-1, siRNA-2 and siRNA-3) and scrambled negative control siRNA (siCtrl), miR-636 mimic and negative control (Genechem, Shanghai, China) were transfected into C666-1 and 5–8F cells, respectively. The full-length of Hsa_circ_0007637 was synthesized (Realgene, Nanjing, China) and cloned into the pcDNA3.1 vector (Genechem, Shanghai, China). Plasmid vectors, including pcDNA3.1-Hsa_circ_0007637 vector and empty vector were, respectively, transfected into 5–8F cells. pcDNA3.1-Hsa_circ_0007637 vector and miR-636 mimic were both used to transfect 5–8F cells. Furthermore, 5–8F cells were cotransfected by pcDNA3.1-Hsa_circ_0007637 vector and TPD52 siRNA (Genechem, Shanghai, China). All the transfection was conducted by applying Lipofectamine 2000 in line with the manuals (Invitrogen, Carlsbad, CA, USA). After 48 h of culture with DMEM (10% FBS) at 37°C, 5% CO2, qRT-PCR was used for transfection efficiency detection.
Cell Counting Kit-8 (CCK-8) Assay
The proliferation of cells was determined by CCK-8 assay. C666-1 and 5–8F cells (1 × 104 cells per well) were seeded into 96-well plates with DMEM (10% FBS) for 24, 48 and 72 h culture at 37°C, 5% CO2. Then, a total of 10 μL CCK-8 solution was added into each well to incubate cells for 2 h at 37°C. The optical density (OD) value was measured using a porous microplate reader (Molecular Devices, Sunnyvale, CA, USA) at 450 nm.
Flow Cytometry
C666-1 and 5–8F cells were harvested after 48 h of transfection, followed by being washed with pro-cooled phosphate buffered saline (PBS). Binding buffer (250 µL) was used to disperse cells. Annexin V/fluorescein isothiocyanate solution (5 µL) and propidium iodide (PI) solution (5 µL) was used for 30 min staining in the dark. The apoptosis was investigated by a flow cytometer (Becton Dickinson, San Jose, CA, USA).
Transwell Experiment
C666-1 and 5–8F cells were digested by trypsin and dispersed in serum-free DMEM (1 × 105 cells/mL). Transwell inserts (8 μm pore size, pre-coated with Matrigel) containing 500 μL of the cell suspension were inserted into 6-well plates. At the bottom of the 6-well plates, 600 μL DMEM containing 10% FBS was added. Cells were kept at 37°C with 5% CO2 for 24 h. After that, the Transwell inserts were removed. The non-invading cells were scraped off using a cotton swab. The invading cells on the lower surface of the chamber were fixed with 4% paraformaldehyde. Then, 0.1% crystal violet was applied to stain cells for 10 min. Invading cells were counted in 5 random fields of view under a microscope (Olympus, Tokyo, Japan).
Wound Healing Assay
C666-1 and 5–8F cells were digested by trypsin and dispersed in DMEM (10% FBS) (1 × 106 cells/mL). The cell suspension was seeded into 6-well plates with 1 mL/well. After 24 h of culture at 37°C, 5% CO2, a wound was made at the bottom of the wells using a sterile pipette tip. The liquid in each well was then discarded. The initial wound width was measured and recorded. Fresh DMEM (10% FBS) was added into each well to incubate cells for 24 h at 37°C, 5% CO2. After discarding the liquid, the final wound width was measured. The relative wound width was calculated by initial wound width/final wound width.
Dual Luciferase Reporter Gene Assay
Through bioinformatics prediction, miR-636, miR-661 and miR-940 might be targets of Hsa_circ_0007637. TPD52 might be a target of miR-636. Dual-luciferase reporter gene assay was performed to verify the binding between the two genes. Briefly, HEK293 cells (1 × 106 cells) were cultured with DMEM (without FBS) in 6-well plates. miR-636 mimic and negative control, miR-661 and negative control, miR-940 and negative control were commercially provided by Genechem (Shanghai, China). They were, respectively, transfected into HEK293 cells using Lipofectamine 2000. pGL3-Hsa_circ_0007637 reporter plasmids containing Hsa_circ_0007637 wild type (WT) and mutant type (Mut) sequence were purchased from Genechem (Shanghai, China). HEK293 cells were then cotransfected by pGL3-Hsa_circ_0007637 reporter plasmids. Moreover, HEK293 cells transfected by miR-636 mimic or negative control were experienced cotransfection with pGL3-TPD52 reporter plasmids containing TPD52 WT or Mut sequence. After cotransfection, HEK293 cells were cultured with DMEM (with 10% FBS) at 37°C, 5% CO2 for 48 h. The luciferase activity was investigated using a Dual Luciferase assay kit (Promega, Madison, WI, USA). The firefly luciferase activity was normalized to Renilla luciferase activity.
RNA Immunoprecipitation (RIP) Experiment
RIP experiment was performed using 5–8F cells by EZ-Magna RIP kit (Millipore, Billerica, MA, USA). The operation was conducted strictly in line with the manuals. Briefly, 5–8F cells transfected by miR-636 mimic and negative control were collected and incubated with RIP lysis buffer. The cell extract with a volume of 100 μL was incubated at 4°C for 6 h with RIP buffer containing magnetic beads conjugated with antibodies against AGO2 or IgG. Thereafter, the beads were washed with a wash buffer. Proteinase K was then added to the complexes for 30 min incubation at 55°C. The immunoprecipitated Hsa_circ_0007637 was evaluated using qRT-PCR.Citation10
RNA Fluorescence in situ Hybridization (FISH) Experiment
The colocalization of Hsa_circ_0007637 and miR-636 in C666-1 and 5–8F cells were detected by RNA FISH experiment. Alexa Fluor 555-labeled Hsa_circ_0007637 probes and Alexa Fluor 488-labeled miR-636 probes were purchased from RiboBio (Guangzhou, China). In line with the manuals, the signals of Hsa_circ_0007637 probes and miR-636 probes were detected using a Fluorescent In Situ Hybridization Kit (RiboBio, Guangzhou, China). The signals were observed under a fluorescence microscope (Eclipse E600; Nikon Corporation, Tokyo, Japan). Hsa_circ_0007637 probes were presented as red fluorescent, while miR-636 probes were exhibited as green fluorescent. The nucleus stained by 4ʹ6’-diamidino-2-phenylindole (DAPI) is presented as blue fluorescence.Citation11
RNA Pull Down Assay
The interaction between miR-636 and TPD52 was verified by RNA pull-down assay. Briefly, 5–8F cells were transfected with biotinylated negative control, biotinylated TPD52 sense or antisense (Genechem, Shanghai, China). Cells were cultured for 48 h at 37°C, 5% CO2. Thereafter, cells were harvested and lysed using a lysis buffer. As described previously, the cell lysates were incubated with M-280 streptavidin magnetic beads (Invitrogen, San Diego, CA, USA).Citation12 The bound RNAs were purified using Trizol. The expression of miR-636 was explored by qRT-PCR.
Western Blot
C666-1 and 5–8F cells were incubated with Lysis Buffer (Solarbio, Beijing, China) on ice to extract total proteins. Protease inhibitor (Solarbio, Beijing, China) was contained in the RIPA Lysis Buffer. The total proteins concentration was researched using a BCA kit (Beyotime, Shanghai, China). An equal amount of each total proteins sample was experienced separation with sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) for 2 h at 100 V. Thereafter, proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane for 2 h at 280 mA. The proteins were treated with 5% skimmed milk for 1 h at room temperature. Primary antibodies were applied to probe the PVDF membrane for 12 h at 4°C. Primary antibodies were rabbit anti-TPD52 (1:1000, ab182578, Abcam, Cambridge, UK) and rabbit anti-β-actin (1:1000, ab8227, Abcam, Cambridge, UK). β-actin was used as the internal control. After that, the proteins were treated by horseradish peroxidase conjugated goat anti-rabbit secondary antibody (1:2000, ab6721, Cambridge, UK) for 2 h at room temperature. Enhanced chemiluminescence kit (Beyotime, Shanghai, China) was used for the visualization of protein bands. The quantification of protein bands was analyzed using the Image J software (National Institutes of Health, Bethesda, Maryland, USA).
In vivo Study
All animal experiments have been approved by the Animal Care and Use Committees of The First Affiliated Hospital of Soochow University. The animal experiments were performed in accordance with relevant guidelines and regulations of the Animal Care and Use Committees at The First Affiliated Hospital of Soochow University. A signed document issued by the Animal Care and Use Committees that granted approval was obtained.
A total of 12 nude mice (5 weeks old) were purchased from Shanghai Experimental Animal Center, Chinese Academy of Sciences (Shanghai, China). Mice were kept in a 12 day/night cycle room at 22°C with free access to water and food. Hsa_circ_0007637 shRNA and negative control were purchased from Genechem (Shanghai, China), followed by being transfected into 5–8F cells using Lipofectamine 2000. Mice were randomly divided into shHsa_circ_0007637 group and shNC group. Then, 5–8F cells (1 × 107 cells) were stably transfected with Hsa_circ_0007637 shRNA and negative control was subcutaneously injected into the right flank of mice. Tumor volume was calculated every 7 days by V = (ab2 × 0.5) (a: long diameter; b: short diameter). On day 28, mice were euthanized to strip tumor tissues. The tumor weight was weighted and immediately stored at −80°C.
qRT-PCR
Total RNAs were extracted from tissues and cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The concentration of each RNA sample was determined by NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA). cDNA templates of miRNAs were synthesized using a miRNA First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) strictly according to the instructions. PrimeScript RT Master Mix (TaKaRa, Dalian, China) was applied for the reverse transcription of mRNA. The qRT-PCR was performed on a LightCycler® 96 System (Roche, Basel, Switzerland) with the following conditions: denaturation at 95°C for 30 s, 40 cycles of amplification at 95°C for 5 s and 60°C for 20 s. The primers were as follows: Hsa_circ_0007637, forward, 5ʹ-AGGAGGCATGGCCAAGATTT-3ʹ, reverse, 5ʹ-TTGATACTAGAGCCGCTGCC-3ʹ. miR-636, forward: 5ʹ-ACACTCCAGCTGGGTGTGC-3ʹ, reverse: 5ʹ-TGGTGTCGTGGAGTCG-3ʹ. miR-661, forward, 5ʹ-ACACTCCAGCTGGGTGCCTGGGTCTCTGGCCT-3ʹ, reverse, 5ʹ-CTCAACTGGTGTCGTGGA-3ʹ. miR-940, forward, 5′-CCTGTCTTACTTTTCCG AAGGAC-3′, reverse, 5′-TTGCTGTATTGTTGCCCATGT-3′. TPD52, forward, 5′-GAGGAAGGAGAAGATGTTGC-3′, reverse, 5′-GCCGAATTCAAGACTTCTCC-3′. U6, forward, 5ʹ-CTCGCTTCGGGCAGCACA-3ʹ, reverse, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ.
β-actin, forward, 5ʹ-ATAGCACAGCCTGGATAGCAACGTAC-3ʹ, reverse, 5ʹ-CACCTTCTACAATGAGCTGCGTGTG-3ʹ. The relative expression of genes was determined by 2−∆∆CT method. U6 was used for the control of miRNAs expression, while β-actin was served as the control for Hsa_circ_0007637 and TPD52 mRNA expression.
Immunohistochemistry (IHC)
TPD52 expression in clinical tissues and Ki67 expression in xenograft tumor tissues were detected by IHC. Tissues were embedded in paraffin to prepare sections (4 μm in thickness). The sections were deparaffinized with xylene, followed by being rehydrated with a series of gradient alcohol (100%, 95%, 90%, 80% and 70%). To remove endogenous peroxidase, 3% H2O2 was used to treat the sections for 10 min. Subsequently, a sodium citrate buffer solution (0.01 M, pH = 6.0) was added to incubate the sections for 3 min under boiling conditions. Bovine serum albumin (Solarbio, China, Beijing) was used to block the section for 30 min at 37°C. Rabbit anti-TPD52 (1:200, ab182578, Abcam, Cambridge, UK) and rabbit anti-Ki67 (1:200, ab15580, Abcam, Cambridge, UK) was used, respectively, to incubate the sections for 12 h at 4°C. The sections were washed with PBS for 3 times. Goat anti-rabbit secondary antibody (1:200, ab6721, Abcam, Cambridge, UK) was added to treat the sections for 30 min at 37°C. The TPD52 and Ki67 positive cells were developed by 3,3′- diaminobenzidine (DAB) staining. Under a microscope (Olympus, Tokyo, Japan), the TPD52 and Ki67 positive cells were presented as brown particles.
Statistical Analysis
All experiments were independently repeated 3 times. SPSS 19.0 (SPSS, Inc., Chicago, IL, USA) was responsible for the data analysis. Data were displayed in the form of mean ± standard deviation. Patients’ survival was evaluated by Kaplan–Meier analysis following Log rank tests. Two-tailed paired Student’s t-test was used for the comparison between the two groups. A one-way analysis of variance followed by Tukey’s post hoc test was conducted for the comparison in multiple groups. The correlation between the two genes was analyzed by Pearson correlation coefficient analysis. P < 0.05 was regarded as a statistically significant difference.
Results
Hsa_circ_0007637 Was Abnormally Overexpressed in NPC Patients and Cells
qRT-PCR displayed significantly higher Hsa_circ_0007637 expression in tumor tissues than that in matched adjacent normal tissues (P < 0.05) (). Relative to patients in low Hsa_circ_0007637 expression group (n = 39), those in high Hsa_circ_0007637 expression group (n = 41) presented markedly lower percent survival (P < 0.05) (). High Hsa_circ_0007637 expression was significantly associated with advanced TNM clinical stage and no keratinizing squamous NPC (P < 0.05) (). In vitro experiment showed that NPC cell lines (C666-1 and 5–8F) exhibited much higher Hsa_circ_0007637 expression than that in nasopharyngeal epithelial cell line (NP460) (P < 0.01) (). Thus, Hsa_circ_0007637 expression was markedly up-regulated in NPC patients and cells.
Table 1 The Correlation Between Hsa_circ_0007637 Expression and NPC Clinical Pathology
Figure 1 Hsa_circ_0007637 was abnormally overexpressed in NPC patients and cells. (A) qRT-PCR indicated that Hsa_circ_0007637 expression was elevated in tumor tissues of NPC patients. (B) Kaplan-Meier survival analysis revealed that high Hsa_circ_0007637 expression was associated with poor percent survival of NPC patients. (C) qRT-PCR illustrated that Hsa_circ_0007637 expression was increased in NPC cell lines (C666-1 and 5–8F) than that in normal nasopharyngeal epithelial cell line (NP460). **P < 0.01 when compared with Hsa_circ_0007637 expression in NP460 cell line. (D) The Hsa_circ_0007637 expression was not affected by RNase R treatment, indicating that Hsa_circ_0007637 had stable circle structure. **P < 0.01 when compared with GAPDH expression in Mock group.
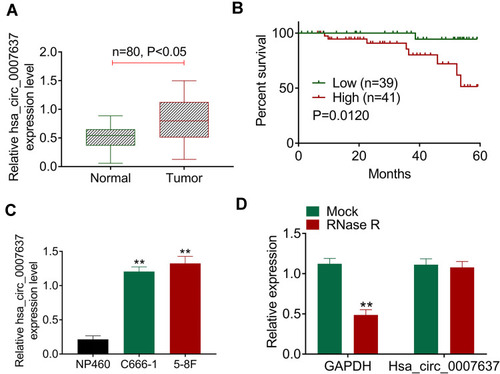
The circle structure stability of Hsa_circ_0007637 was assessed by RNase R treatment. The results are shown in . Obviously, RNase R treatment remarkably reduced GAPDH expression (P < 0.01). However, Hsa_circ_0007637 expression was not obviously affected by RNase R treatment. Hence, the circle structure of Hsa_circ_0007637 was not destroyed by RNase R treatment, indicating that Hsa_circ_0007637 had stable circle structure.
Hsa_circ_0007637 Knockdown Decreased Proliferation, Invasion, Migration and Increased Apoptosis of NPC Cells
From , C666-1 and 5–8F cells of siHsa_circ_0007637-1 group, siHsa_circ_0007637-2 group and siHsa_circ_0007637-3 group showed significantly lower Hsa_circ_0007637 expression than that of siCtrl group (P < 0.01). The transfection efficiency of Hsa_circ_0007637 siRNA-1 was higher than Hsa_circ_0007637 siRNA-2 and Hsa_circ_0007637 siRNA-3. Thus, C666-1 and 5–8F cells of siHsa_circ_0007637-1 group (renamed as siHsa_circ_0007637 group in the following study) were used to research the Hsa_circ_0007637 function on C666-1 and 5–8F cells malignant phenotype.
Figure 2 Hsa_circ_0007637 knockdown decreased proliferation, invasion, migration and increased apoptosis of NPC cells. (A) Hsa_circ_0007637 expression in C666-1 and 5–8F cells was successfully silenced by transfection. **P < 0.01 when relative to siCtrl group. (B) CCK-8 assay indicated that Hsa_circ_0007637 knockdown decreased proliferation of NPC cells. **P < 0.01. (C) Flow cytometry revealed that Hsa_circ_0007637 knockdown enhanced apoptosis of NPC cells. **P < 0.01 when relative to siCtrl group. (D) Transwell experiment illustrated that Hsa_circ_0007637 knockdown reduced invasion of NPC cells. **P < 0.01 when relative to siCtrl group. (E) Wound healing assay demonstrated that Hsa_circ_0007637 knockdown declined migration of NPC cells. **P < 0.01 when relative to siCtrl group.
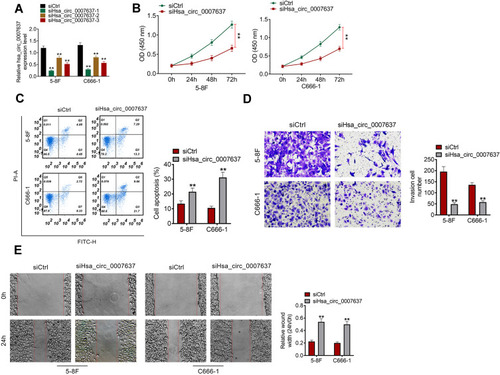
CCK-8 assay, flow cytometry, transwell experiment and wound healing assay were performed, respectively, to detect C666-1 and 5–8F cells proliferation, apoptosis, invasion and migration abilities. Relative to siCtrl group, C666-1 and 5–8F cells of siHsa_circ_0007637 group exhibited dramatically lower OD value, higher apoptosis, lower invasion cell number and higher relative wound width (all P < 0.01) (–). Therefore, Hsa_circ_0007637 knockdown decreased proliferation, invasion, migration and increased apoptosis of NPC cells.
miR-636 Was a Target Gene of Hsa_circ_0007637
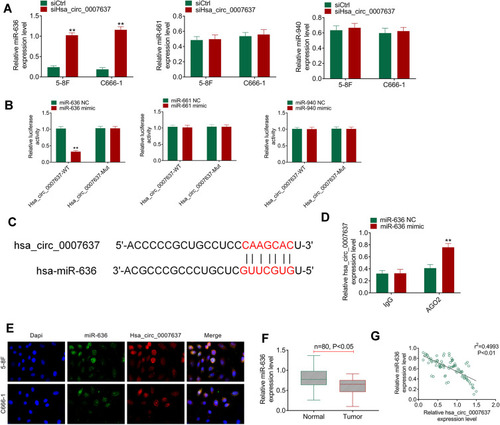
Through bioinformatics prediction, it was found that miR-636, miR-661 and miR-940 might be potential targets of Hsa_circ_0007637. The expression of miR-636, miR-661 and miR-940 in the transfected C666-1 and 5–8F cells was detected. Compared with siCtrl group, C666-1 and 5–8F cells of siHsa_circ_0007637 group had dramatically higher miR-636 expression (P < 0.01). However, the difference in miR-661 and miR-940 expression between siCtrl group and siHsa_circ_0007637 group was not obvious (). Dual-luciferase reporter gene assay showed that, relative to miR-636 NC group, HEK293 cells of miR-636 mimic group exhibited significantly lower Hsa_circ_0007637-WT luciferase activity (P < 0.01). The difference in Hsa_circ_0007637-Mut luciferase activity was not obviously changed between miR-636 NC group and miR-636 mimic group. When compared with miR-661 NC group, the transfection of miR-661 mimic did not obviously change the luciferase activity of Hsa_circ_0007637-WT and Hsa_circ_0007637-Mut. Similar results were observed in HEK293 cells of miR-940 NC group and miR-940 mimic group (). The binding site of Hsa_circ_0007637 and miR-636 is shown in . RIP experiment by 5–8F cells showed that Hsa_circ_0007637 was obviously precipitated by AGO2 antibody. Relative to miR-636 NC group, Hsa_circ_0007637 expression was distinctly increased in miR-636 mimic group (P < 0.01) (). RNA FISH experiment displayed that Hsa_circ_0007637 and miR-636 were mainly colocalized in the cytoplasm (). In tumor tissues of NPC patients, miR-636 was lower expressed than that in adjacent normal tissues (P < 0.05) (). The Hsa_circ_0007637 and miR-636 expression presented a significant negative correlation in tumor tissues of NPC patients (P < 0.01) (). All of these data identified that miR-636 was a target gene of Hsa_circ_0007637.
Figure 3 miR-636 was a target gene of Hsa_circ_0007637. (A) Hsa_circ_0007637 knockdown markedly increased miR-636 expression in NPC cells, but had no obvious effect on miR-661 and miR-940 expression. **P < 0.01 relative to siCtrl group. (B) Dual luciferase reporter gene assay indicated that miR-636 was a target gene of Hsa_circ_0007637. **P < 0.01 relative to miR-636 NC group. (C) The binding site of Hsa_circ_0007637 and miR-636 was shown. (D) RIP experiment revealed that Hsa_circ_0007637 could be precipitated by miR-636. **P < 0.01 relative to miR-636 NC group. (E) RNA FISH experiment displayed that Hsa_circ_0007637 and miR-636 was mainly colocalized in the cytoplasm. (F) miR-636 was lower expressed in tumor tissues of NPC patients. (G) The Hsa_circ_0007637 and miR-636 expression presented a significant negative correlation in tumor tissues of NPC patients.
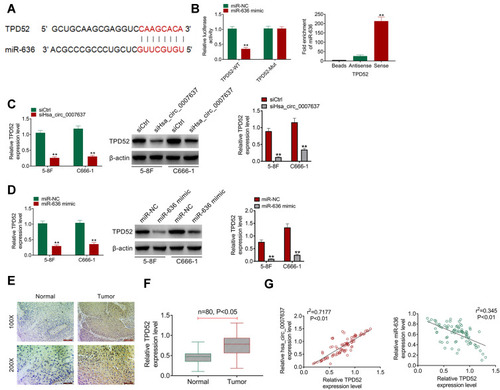
TPD52 Was Directly Inhibited by miR-636
According to bioinformatics prediction, TPD52 had binding site of miR-636 (). Dual-luciferase reporter gene assay exhibited that miR-636 overexpression had no obvious effects on TPD52-Mut luciferase activity. However, significantly reduced TPD52-WT luciferase activity was observed after miR-636 being overexpressed in HEK293 cells (P < 0.01). RNA pull-down assay presented that miR-636 was remarkably pulled down by TPD52 (P < 0.01) (). qRT-PCR and Western blot showed that C666-1 and 5–8F cells of siHsa_circ_0007637 group had a much lower TPD52 mRNA and protein expression (P < 0.01) (). Simultaneously, C666-1 and 5–8F cells of miR-636 mimic group presented markedly lower TPD52 mRNA and protein expression than that of miR-636 NC group (P < 0.01) (). In order to research TPD52 expression, IHC and qRT-PCR was performed on clinical tissues of NPC patients. IHC result showed more TPD52 positive cells in tumor tissues than those in adjacent normal tissues (). qRT-PCR displayed significantly higher TPD52 mRNA expression in tumor tissues than that in adjacent normal tissues (P < 0.05) (). In tumor tissues of NPC patients, Hsa_circ_0007637 and TPD52 mRNA expression showed a significant positive correlation (P < 0.01). miR-636 and TPD52 mRNA expression exhibited a significant negative correlation (P < 0.01) (). Thus, TPD52 was confirmed as a target gene of miR-636.
Figure 4 TPD52 was directly inhibited by miR-636. (A) The binding site of TPD52 and miR-636 was shown. (B) Dual luciferase reporter gene assay revealed that TPD52 was a target gene of miR-636. RNA pull down assay indicated that miR-636 was remarkably pulled down by TPD52. **P < 0.01 compared with miR-636 NC group or Antisense group. (C) qRT-PCR and Western blot demonstrated that Hsa_circ_0007637 knockdown reduced TPD52 mRNA and protein expression in NPC cells. **P < 0.01 compared with siCtrl group. (D) miR-636 overexpression declined TPD52 mRNA and protein expression in NPC cells. **P < 0.01 compared with miR-636 NC group. (E) IHC showed more TPD52 positive cells in tumor tissues than that in adjacent normal tissues. (F) qRT-PCR displayed that TPD52 mRNA expression was significantly elevated in tumor tissues of NPC patients. (G) In tumor tissues of NPC patients, TPD52 mRNA expression was positively correlated with Hsa_circ_0007637, but was negatively correlated with miR-636.
Hsa_circ_0007637 Promoted NPC Progression by Sponging miR-636/TPD52 Axis
Rescue experiment was performed by cotransfection of 5–8F cells. Relative to Ctrl group, much higher Hsa_circ_0007637 expression in Hsa_circ_0007637 group, Hsa_circ_0007637 + miR-636 mimic group and Hsa_circ_0007637 + siTPD52 group was found (P < 0.01). Much lower miR-636 expression was observed in 5–8F cells of Hsa_circ_0007637 group and Hsa_circ_0007637 + siTPD52 group when compared with Ctrl group (P < 0.01). Furthermore, TPD52 expression in Hsa_circ_0007637 group was significantly increased than that in Ctrl group (P < 0.01). Conversely, obviously decreased TPD52 expression was observed in 5–8F cells of Hsa_circ_0007637 + miR-636 mimic group and Hsa_circ_0007637 + siTPD52 group when relative to Hsa_circ_0007637 group (P < 0.01) (). Thus, 5–8F cells were successfully transfected.
Figure 5 Hsa_circ_0007637 promoted NPC progression by sponging miR-636/TPD52 axis. (A) Hsa_circ_0007637, miR-636 and TPD52 expression in 5–8F cells was successfully regulated by transfection. **P < 0.01 relative to Ctrl group. (B) CCK-8 assay revealed that Hsa_circ_0007637 enhanced 5–8F cells proliferation by sponging miR-636/TPD52 axis. **P < 0.01. (C) Flow cytometry demonstrated that Hsa_circ_0007637 weakened 5–8F cells apoptosis by sponging miR-636/TPD52 axis. **P < 0.01. (D) Wound healing assay indicated that Hsa_circ_0007637 promoted 5–8F cells migration by sponging miR-636/TPD52 axis. **P < 0.01. (E) Transwell experiment illustrated that Hsa_circ_0007637 facilitated 5–8F cells invasion by sponging miR-636/TPD52 axis. **P < 0.01.
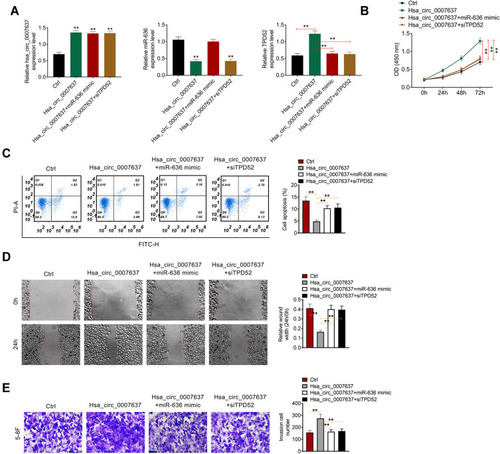
CCK-8 assay, flow cytometry, wound healing assay and transwell experiment were used to research 5–8F cells proliferation, apoptosis, migration and invasion respectively. As a result, 5–8F cells of Hsa_circ_0007637 group presented dramatically higher OD value, lower apoptosis and relative wound width, and higher invasion cell number than that of Ctrl group (P < 0.01). In comparison with Hsa_circ_0007637 group, 5–8F cells of Hsa_circ_0007637 + miR-636 mimic group and Hsa_circ_0007637 + siTPD52 group showed remarkably lower OD value, higher apoptosis and relative wound width, and lower invasion cell number (P < 0.01) (–). These results indicated that Hsa_circ_0007637 might promote NPC progression by sponging miR-636/TPD52 axis.
Hsa_circ_0007637 Knockdown Suppressed NPC Cells Growth in vivo
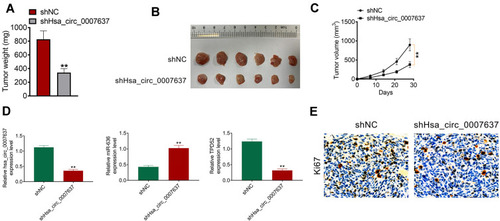
The function of Hsa_circ_0007637 on NPC cells growth in vivo was explored by xenograft tumor experiment. On day 28 after subcutaneous injection, mice of shHsa_circ_0007637 group had a much lower tumor weight and volume than that of shNC group (P < 0.01) (–). qRT-PCR was used to research Hsa_circ_0007637, miR-636 and TPD52 expression in xenograft tumors. Relative to shNC group, shHsa_circ_0007637 group exhibited remarkably lower Hsa_circ_0007637 and TPD52 expression, and higher miR-636 expression (P < 0.01) (). IHC displayed that less Ki67 positive cells in xenograft tumors of shHsa_circ_0007637 group were shown when matched with shNC group ().
Figure 6 Hsa_circ_0007637 knockdown suppressed NPC cells growth in vivo. (A) On day 28 after subcutaneous injection, mice of shHsa_circ_0007637 group had much lower tumor weight than that of shNC group. **P < 0.01 relative to shNC group. (B) The xenograft tumors were obtained after 28 days of subcutaneous injection. (C) Mice of shHsa_circ_0007637 group had much lower volume weight than that of shNC group. **P < 0.01. (D) qRT-PCR indicated that Hsa_circ_0007637 knockdown reduced Hsa_circ_0007637 and TPD52expression, but elevated miR-636 expression in xenograft tumors. **P < 0.01 relative to shNC group. (E) IHC revealed that Hsa_circ_0007637 knockdown decreased Ki67 expression in xenograft tumors.
Discussion
circRNAs have attracted increasing attention in recent years. It is not easily degraded by nucleic acid exonuclease due to the special closed-loop structure. Hence, circRNAs are more stable than long non-coding RNAs or microRNAs. It becomes a potential target and biomarker for disease treatment and diagnosis.Citation13 In this study, Hsa_circ_0007637 had stably closed-loop structure, which could not be easily digested by RNase R. More importantly, Hsa_circ_0007637 expression was abnormally up-regulated in NPC tissues and cells. High Hsa_circ_0007637 expression was associated with poor outcomes in NPC patients. Hsa_circ_0007637 knockdown weakened NPC cells malignant phenotype in vitro and growth in vivo. This was the first time that the function of Hsa_circ_0007637 in NPC progression was studied in detail.
NPC has the characteristics of high malignancy and metastasis rate. Target therapy for NPC has become a research hotspot in recent years. Accumulating evidence revealed that circRNAs were involved in the pathogenesis of NPC. For example, hsa_circ_0081534 might be a novel target for NPC target treatment because hsa_circ_0081534 inhibition reduced NPC cells growth both in vitro and in vivo by sponging miR-508-5p.Citation14 Similarly, Circ-ZNF609 was found to be highly expressed in NPC. circ-ZNF609 silencing aberrantly restrained proliferation and accelerated apoptosis of NPC cells via sponging miR-188.Citation15 Moreover, NPC patients expressed more circ_0033074 in tumor tissues than that in adjacent normal tissues. In vitro study indicated that the inhibition of circ_0033074 remarkably suppressed NPC cells proliferation and invasion.Citation16 Simultaneously, hsa_circ_0028007, hsa_circ_0066755 and circ-ZNF609 were demonstrated to be oncogene of NPC, which facilitated the malignant development of NPC through sponging its downstream miRNAs.Citation17–Citation19 Guo et alCitation20 identified that the lower expressed circPSMC3 was associated with poor prognosis in NPC patients. Meanwhile, circTGFBR2 was discovered to be down-regulated in NPC tissues. The high circTGFBR2 expression in NPC patients was related to favorable prognosis of patients.Citation21 All of these studies revealed the important regulatory roles of circRNAs in NPC progression. In terms of Hsa_circ_0007637, it has rarely been studied in human diseases. Only a recent study indicated that Hsa_circ_0007637 was differentially expressed in NPC.Citation9 Unfortunately, the study did not explore the detailed function of Hsa_circ_0007637 in regulating NPC development. Our study firstly demonstrated that Hsa_circ_0007637 acted as an oncogene in NPC.
Usually, circRNAs participate in the regulation of human tumors by acting as competing endogenous RNAs. It could regulate mRNA expression by sponging miRNAs.Citation21 In this article, it was identified that miR-636 was a target gene of Hsa_circ_0007637. Hsa_circ_0007637 facilitated NPC progression by enhancing TPD52 expression via sponging miR-636. At present, studies focused on miR-636 regulation of NPC development have not been found. According to a previous study, miR-636 has been found to be down-regulated in cervical cancer. By overexpression of miR-636, cervical cancer cells proliferation was suppressed and apoptosis was enhanced. The opposite effect was discovered by down-regulation of miR-636.Citation22 Chen et alCitation23 detected the expression of miR-636 in the plasma of urothelial carcinoma patients. miR-636 expression was markedly different between urothelial carcinoma patients and non-urothelial carcinoma patients. However, the study did not continue in-depth experiments to verify the influence of miR-636 on urothelial carcinoma development. In some other human tumors, the expression of miR-636 has been researched to be regulated by several circRNAs, such as circRNA_100876 and CircPTPRA.Citation24,Citation25 Our study firstly detected the expression of miR-636 in NPC patients. It was shown that miR-636 was lower expressed in tumor tissues of NPC patients. RNA FISH experiment indicated that miR-636 and Hsa_circ_0007637 were mainly colocalized in the cytoplasm. Rescue experiment illustrated that miR-636 up-regulation reversed the promoting effect of Hsa_circ_0007637 on NPC cells malignant phenotype in vitro.
TPD52 was identified as a target gene of miR-636 in this study. Hsa_circ_0007637 enhanced NPC progression by elevating TPD52 expression via sponging miR-636. TPD52 is located at chromosome 8q21, which is frequently amplified in several human tumors.Citation26 The high TPD52 expression has been explored to be associated with worse progression of pancreatic cancer, breast cancer and colorectal cancer, etc.Citation27–Citation29 Yin et alCitation30 reported that TPD52 was involved in the regulation of NPC progression. miR-449b-5p suppressed NPC cells malignant phenotype in vitro by inhibiting the expression TPD52. In particular, Zhao et alCitation31 illustrated that TPD52 knockdown remarkably restrained NPC cells proliferation, invasion and migration. Our paper demonstrated that TPD52 was highly expressed in tumor tissues of NPC patients. TPD52 knockdown reversed the promoting effect of Hsa_circ_0007637 on NPC cells malignant phenotype in vitro. Previous research has investigated that TPD52 knockdown could suppress tumor cells proliferation, migration, invasion and induce tumor cells apoptosis by inhibiting the activity of Akt pathway.Citation27 However, it has been fully identified that the inhibition of Akt pathway activity was conducive to the suppression of NPC progression.Citation32,Citation33 Thus, we speculated that TPD52 knockdown might be involved in the NPC progression inhibition by suppressing the activity of Akt pathway.
Conclusion
In summary, this study concluded, for the first time, that Hsa_circ_0007637 was highly expressed in NPC patients and cells. High Hsa_circ_0007637 expression was associated with poor prognosis of NPC patients. Hsa_circ_0007637 knockdown suppressed NPC progression both in vitro and in vivo. Mechanism research indicated that Hsa_circ_0007637 might facilitate NPC progression by enhancing TPD52 expression via sponging miR-636. Therefore, Hsa_circ_0007637 might be used as a promising candidate for NPC target treatment.
Disclosure
The authors report no conflicts of interest in this work.
References
- Yin L, Chen J, Ma C, et al. Hsa_circ_0046263 functions as a ceRNA to promote nasopharyngeal carcinoma progression by upregulating IGFBP3. Cell Death Dis. 2020;11(7):562. doi:10.1038/s41419-020-02785-332703944
- Lam WKJ, Chan JYK. Recent advances in the management of nasopharyngeal carcinoma. F1000Res. 2018;7:F1000 Faculty Rev–1829.
- Guo R, Mao YP, Tang LL, et al. The evolution of nasopharyngeal carcinoma staging. Br J Radiol. 2019;92(1102):20190244. doi:10.1259/bjr.2019024431298937
- Ke KJ, Wang CY, Li P, et al. Hsa_circ_001193 regulates proliferation and apoptosis of nasopharyngeal carcinoma cells through targeting mir-496. Eur Rev Med Pharmacol Sci. 2020;24(6):3069–3076.32271424
- Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. doi:10.1186/s12943-018-0827-829626935
- Yang X, Ye T, Liu H, et al. Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer. Mol Cancer. 2021;20(1):4. doi:10.1186/s12943-020-01300-833397425
- Fang Q, Liu H, Zhou A, et al. Circ_0046599 promotes the development of hepatocellular carcinoma by regulating the miR-1258/RPN2 network. Cancer Manag Res. 2020;12:6849–6860. doi:10.2147/CMAR.S25351032801909
- He Y, Wang Y, Liu L, et al. Circular RNA circ_0006282 contributes to the progression of gastric cancer by sponging miR-155 to upregulate the expression of FBXO22. Onco Targets Ther. 2020;13:1001–1010. doi:10.2147/OTT.S22821632099403
- Wu H, Liu Y, Duan H, et al. Identification of differentially expressed circular RNAs in human nasopharyngeal carcinoma. Cancer Biomark. 2020;29(4):483–492. doi:10.3233/CBM-20173132865182
- Chen QN, Chen X, Chen ZY, et al. Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with EZH2 and repressing IL24 expression. Mol Cancer. 2017;16(1):17. doi:10.1186/s12943-017-0581-328109288
- Wu Y, Xie Z, Chen J, et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression. Mol Cancer. 2019;18(1):73. doi:10.1186/s12943-019-1007-130940151
- Xiao Z, Qiu Y, Lin Y, et al. Blocking lncRNA H19-miR-19a-Id2 axis attenuates hypoxia/ischemia induced neuronal injury. Aging. 2019;11(11):3585–3600. doi:10.18632/aging.10199931170091
- Duan ZN, Dong CG, Liu JH. Circ-ABCB10 promotes growth and metastasis of nasopharyngeal carcinoma by upregulating ROCK1. Eur Rev Med Pharmacol Sci. 2020;24(23):12208–12215.33336739
- Li S, Wang Q. Hsa_circ_0081534 increases the proliferation and invasion of nasopharyngeal carcinoma cells through regulating the miR-508-5p/FN1 axis. Aging. 2020;12(20):20645–20657. doi:10.18632/aging.10396333082297
- Li M, Li Y, Yu M. CircRNA ZNF609 knockdown suppresses cell growth via modulating miR-188/ELF2 axis in nasopharyngeal carcinoma. Onco Targets Ther. 2020;13:2399–2409. doi:10.2147/OTT.S23423032273713
- Liu R, Zhou M, Zhang P, et al. Cell proliferation and invasion is promoted by circSERPINA3 in nasopharyngeal carcinoma by regulating miR-944/MDM2 axis. J Cancer. 2020;11(13):3910–3918. doi:10.7150/jca.4279932328195
- Qiongna D, Jiafeng Z, Yalin H, et al. Implication of hsa_circ_0028007 in reinforcing migration, invasion, and chemo-tolerance of nasopharyngeal carcinoma cells. J Clin Lab Anal. 2020;34(9):e23409. doi:10.1002/jcla.2340932524687
- Wang J, Kong J, Nie Z, et al. Circular RNA Hsa_circ_0066755 as an oncogene via sponging miR-651 and as a promising diagnostic biomarker for nasopharyngeal carcinoma. Int J Med Sci. 2020;17(11):1499–1507. doi:10.7150/ijms.4702432669952
- Zhu L, Liu Y, Yang Y, et al. CircRNA ZNF609 promotes growth and metastasis of nasopharyngeal carcinoma by competing with microRNA-150-5p. Eur Rev Med Pharmacol Sci. 2019;23(7):2817–2826.31002133
- Guo DQ, Liu F, Zhang L, et al. CircPSMC3 inhibits cell proliferation and induces cell apoptosis in nasopharyngeal carcinoma by downregulating ROCK1. Eur Rev Med Pharmacol Sci. 2020;24(20):10306.33155260
- Li W, Lu H, Wang H, et al. Circular RNA TGFBR2 acts as a ceRNA to suppress nasopharyngeal carcinoma progression by sponging miR-107. Cancer Lett. 2021;499:301–313. doi:10.1016/j.canlet.2020.11.00133160003
- Hu QL, Xu ZP, Lan YF, et al. miR-636 represses cell survival by targeting CDK6/Bcl-2 in cervical cancer. Kaohsiung J Med Sci. 2020;36(5):328–335. doi:10.1002/kjm2.1218131889428
- Chen CL, Lin CH, Li AL, et al. Plasma miRNA profile is a biomarker associated with urothelial carcinoma in chronic hemodialysis patients. Am J Physiol Renal Physiol. 2019;316(6):F1094–F1102. doi:10.1152/ajprenal.00014.201930892932
- Song J, Shi W, Gao Z, et al. Downregulation of circRNA_100876 inhibited progression of NSCLC in vitro via targeting miR-636. Technol Cancer Res Treat. 2020;19:1533033820951817. doi:10.1177/153303382095181733030101
- He Q, Huang L, Yan D, et al. CircPTPRA acts as a tumor suppressor in bladder cancer by sponging miR-636 and upregulating KLF9. Aging. 2019;11(23):11314–11328. doi:10.18632/aging.10253031821171
- Wang Y, Chen CL, Pan QZ, et al. Decreased TPD52 expression is associated with poor prognosis in primary hepatocellular carcinoma. Oncotarget. 2016;7(5):6323–6334. doi:10.18632/oncotarget.631926575170
- Wang Z, Li Y, Fan L, et al. Silencing of TPD52 inhibits proliferation, migration, invasion but induces apoptosis of pancreatic cancer cells by deactivating Akt pathway. Neoplasma. 2020;67(2):277–285. doi:10.4149/neo_2019_190404N29531847526
- Roslan N, Bièche I, Bright RK, et al. TPD52 represents a survival factor in ERBB2-amplified breast cancer cells. Mol Carcinog. 2014;53(10):807–819. doi:10.1002/mc.2203823661506
- Li J, Li Y, Liu H, et al. The four-transmembrane protein MAL2 and tumor protein D52 (TPD52) are highly expressed in colorectal cancer and correlated with poor prognosis. PLoS One. 2017;12(5):e0178515. doi:10.1371/journal.pone.017851528562687
- Yin W, Shi L, Mao Y. MicroRNA-449b-5p suppresses cell proliferation, migration and invasion by targeting TPD52 in nasopharyngeal carcinoma. J Biochem. 2019;166(5):433–440. doi:10.1093/jb/mvz05731350893
- Zhao X, Chu J. MicroRNA-379 suppresses cell proliferation, migration and invasion in nasopharyngeal carcinoma by targeting tumor protein D52. Exp Ther Med. 2018;16(2):1232–1240.30116374
- Gao R, Feng Q, Tan G. microRNA-613 exerts anti-angiogenic effect on nasopharyngeal carcinoma cells through inactivating the AKT signaling pathway by down-regulating FN1. Biosci Rep. 2019;39(7):BSR20182196. doi:10.1042/BSR2018219631189740
- Zuo WN, Zhu H, Li LP, et al. MiR-155 promotes proliferation and inhibits apoptosis of nasopharyngeal carcinoma cells through targeting PTEN-PI3K/AKT pathway. Eur Rev Med Pharmacol Sci. 2019;23(18):7935–7942.31599418
