Abstract
Purpose
Long non-coding RNAs (lncRNAs) have diverse roles in modulating gene expression on both transcriptional and translational levels, but their involvement in osteosarcoma (OS) metastasis remains unknown.
Patients and Methods
Transcriptional and clinical data were downloaded from TARGET datasets. A total of seven lncRNAs screened by univariate cox regression, lasso regression, and multivariate cox regression analysis were used to establish the OS metastasis model. The area under the receiver operating characteristic curve (AUC) was used to evaluate the model.
Results
The established model showed exceptional predictive performance (1 year: AUC = 0.92, 95% Cl = 0.83–0.99; 3 years: AUC = 0.87, 95% Cl = 0.79–0.96; 5 years: AUC = 0.86, 95% Cl = 0.76–0.96). Patients in the high group had a poor survival outcome than those in the low group (p < 0.0001). GSEA analysis revealed that “NOTCH_SIGNALING” and “WNT_BETA_CATENIN_SIGNALING” were significantly enriched and that resting dendritic cells were associated with AL512422.1, AL357507.1, and AC006033.2 (p < 0.05).
Conclusion
Based on seven prognosis-related lncRNAs, we constructed a novel model with high reliability and accuracy for predicting metastasis in OS patients.
Introduction
OS is the most common primary malignant bone tumorCitation1 with a bimodal age distribution. The first peak occurs during adolescence, while the second peak occurs later in life.Citation2–Citation6 With the advancements of treatments, surgery and intensive multiagent chemotherapy became the mainstream strategies.Citation7–Citation9 The high incidence of metastasis in OS, particularly lung metastasis, results in a poor prognosis.Citation10–Citation12 The 5-year overall survival rate for non-metastatic OS is about 75%; while it is only 20% in metastatic patients.Citation13,Citation14 Thus, developing biomarkers for early detection and treatment is essential.
LncRNA is a non-coding RNA ranging length from 200 nucleotides (nt) to more than 100 kilobases (kb).Citation15 LncRNA modulates gene expression on both transcriptional and translational levels, including chromosomal rearrangement, histone modification, alternative splicing formation, and RNA stabilization.Citation16 LncRNA has been found to play a role in tumor differentiation, invasion, and metastasis.Citation17–Citation21 LncRNA is implicated in the onset and progression of cancer metastasis. MALAT1, for example, primarily regulates lung cancer metastasis through epithelial-mesenchymal transition (EMT).Citation22,Citation23 It has also been shown to promote metastasis in bladder cancer by activating Wnt signaling.Citation24,Citation25 HOTAIR promotes pancreatic cancer metastasis by suppressing interferon-related genes.Citation26 MEG3 plays a decisive role in meningioma and gastric cancer by regulating autophagy and DNA repair.Citation27,Citation28 However, the role of lncRNA in OS remains unclear, and the lncRNA-based prediction model is currently lacking.
Advances in high-throughput technologies allow researchers to identify biomarkers that can be used to construct prediction models for personalized cancer treatment. In this study, we presented a new risk-score model for predicting the metastasis of OS patients based on lncRNAs. In addition, we would detect the critical biological process, immune-related cells, and immune checkpoint between high and low risk group.
Materials and Methods
Target Data and Processing
The clinical information and expression profiles, including mRNA and lncRNA expression profiles, were downloaded from the TARGET datasets (https://ocg.cancer.gov/programs/target). All datasets were subjected to standardized data preprocessing. In this study, a total of 22 metastasis and 66 primary OS tissue-related data were analyzed. Data pre-processing was performed using RStudio and included background correction, normalization, and batch effect adjustment.
Feature Selection and Construction of Prognostic Model
The Limma R package was used to compare differential gene expression of lncRNA and mRNA in 22 metastasis and 66 primary OS patients. P-value<0.05 and |log 2 (fold change [FC]) | >1 were set as the threshold criteria. Differentially expressed lncRNA in this study, were screened by univariate cox regression, least absolute shrinkage, and selection operator (LASSO) regression, and multivariate cox regression analysis in survival and glmnet R packages. The lncRNA-based prognosis risk score was established using a linear formula with the expression level multiplied regression model (β). The median risk score was used to categorize patients into the high-risk group and low-risk group. The survival and survival ROC R packages were used to plot Kaplan–Meier (KM) survival and receiver operating characteristic (ROC) curves.
Scale-Free Co-Expression Network and Enrichment Analysis
Prognosis-related lncRNAs and metastasis-related mRNA were used to reconstruct the scale-free co-expression network. A total of 24 metastasis-related mRNAs associated with 7 metastasis-related lncRNAs (correlation coefficient R>0.3, p<0.001) were selected. Cytoscape software was used for network visualization. Using DAVID, we investigated the interactions between 24 metastasis-related mRNA and transcription factors (TF). GO pathway enrichment analyses were performed using the R package clusterProfiler. P<0.05 was considered as used as the cut-off for statistical significance.
Gene Set Enrichment Analysis and Gene Set Variation Analysis
GSEA2.2.4 software was used to perform gene set enrichment analysis (GSEA) and generate an enrichment score. The gene sets used for the enrichment analysis were downloaded from the Molecular Signatures Database (MsigDB). After 1000 permutations, gene sets with a false discovery rate (FDR) less than 0.05 were reported to be significantly enriched. Gene Set Variation Analysis (GSVA) is a method for estimating the variation of gene set enrichment using samples from an expression data set. GSVA was performed using the Hallmark gene set in the GSVA package (from R Project 3.5.1) of R software with default parameters.
Infiltrating Immune Cells
The CIBERSORT algorithm (http://cibersort.stanford.edu), a deconvolution algorithm based on gene expression profiles, was used to evaluate the 22 different types of tumor-infiltrating immune cells in the metastatic and primary OS group. CIBERSORT can assess the composition of tumor-infiltrating immune cells, by using P-value as a criterion for measuring confidence in findings. The relationship between 7 prognosis-related lncRNAs and tumor-infiltrating immune cells was also investigated.
Cell Lines and qPCR
Highly metastasis human OS cell lines (MNNG) and weakly metastasis human OS cell lines (U2S2, SAOS-2, SJSA-1, and HOS) were purchased from iCell (Shanghai, China). Trizol reagent (Invitrogen Life Technologies) was used to isolate total RNA. Approximately 2 μg RNA was reverse-transcribed into complementary DNA following the manufacturer’s instructions and normalized using glyceraldehyde 3-phosphate dehydrogenase mRNA levels. Reverse-transcription quantitative PCR assay with PrimeScript RT Master Mix (Takara) was performed on a QuantStudio6 Flex real-time PCR system (Applied Biosystems). The following were the PCR amplification conditions: 2 min at 95°C for one cycle, followed by denaturation for 15 sec at 95°C, and extension for 60 sec at 60°C for 38 cycles. Table S1 presents the primers used.
Result
Differential Gene and lncRNA Screening
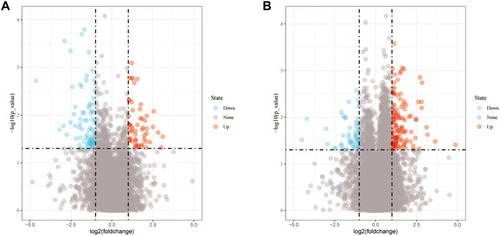
This study included 22 metastatic and 66 primary OS samples. The Limma R package was used to select differentially expressed genes and lncRNA. As a result, 135 differentially expressed lncRNAs were identified, including 79 upregulated and 56 downregulated lncRNAs (). In addition, there were 171 differentially expressed genes ().
Construction of Prognosis Assessment Model
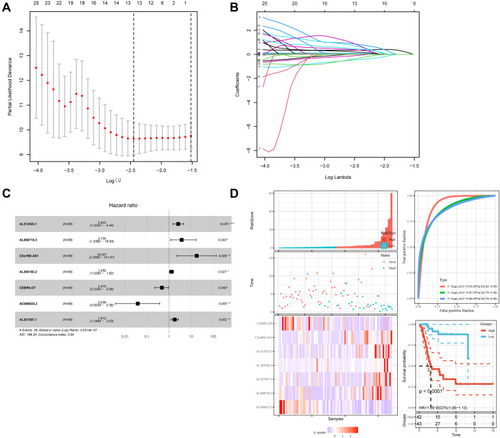
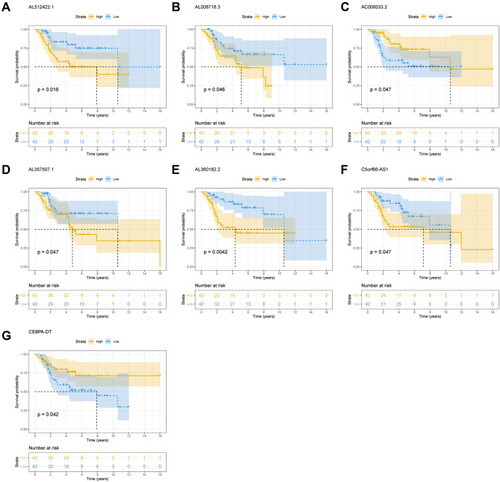
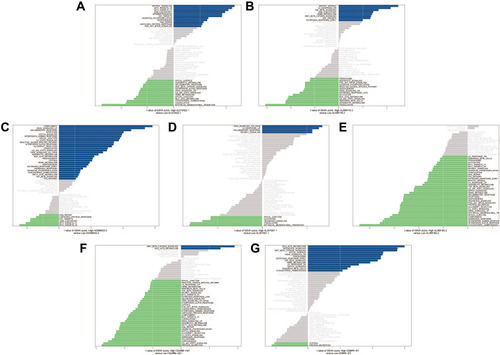
The findings from the univariate Cox regression analysis of 135 lncRNAs were used in LASSO regression to identify potential markers. Multivariate cox regression analysis identified seven prognosis-related lncRNAs (–), including AL512422.1, AL008718.3, C5orf66-AS1, AL360182.2, CEBPA-DT, AC006033.2, and AL357507.1. All of these lncRNAs were used to establish the prognosis model, based on the following formula, y= 2.603*AL512422.1+3.735*AL008718.3+18.047*C5orf66-AS1+1.292*AL360182.2+0.475*CEBPA-DT+0.038*AC006033.2+1.812*AL357507.1. The patients were divided into high-risk and low-risk groups based on the median risk score calculated using the above formula. depicts the expression of seven prognosis-related lncRNAs in high and low-risk groups. Moreover, the model had higher prediction ability, with all area under the curve (AUC) values >0.8 (1 year: AUC = 0.92, 95% Cl = 0.83–0.99; 3 years: AUC = 0.87, 95% Cl = 0.79–0.96; and 5 years: AUC = 0.86, 95% Cl = 0.76–0.96) (). According to the KM survival curves (), the high-risk group had significantly poorer overall survival time than the low-risk group (p<0.0001, HR=1.09, 95% CI = 1.06–1.13). In addition, the KM survival curves indicated that five lncRNAs, including AL512422.1, AL008718.3, C5orf66-AS1, AL360182.2, and AL357507.1, were associated with poor overall survival. CEBPA-DT and AC006033.2, on the other hand, were associated with improved survival (–).
Co‑Expression Network and Enrichment Analysis
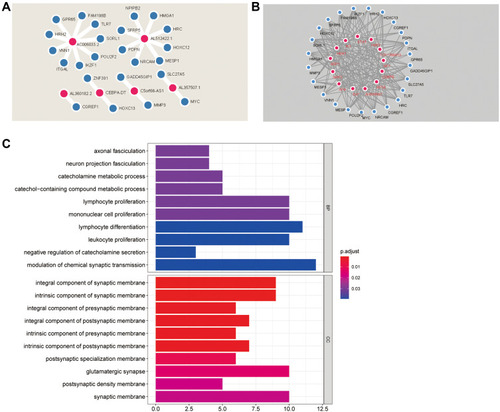
To explore the mRNAs associated with the model-related lncRNAs and their potential TF, we integrated the lncRNA and gene data to construct a co-expression network. The metastasis-related genes that were closely associated with the seven prognosis-related lncRNAs were selected (R>0.3, p<0.001). A total of 6 prognosis-related lncRNAs and 24 metastasis-related genes were used to construct the co-expression network (). AC006033.2 and AL512422.1 were the most important nodes. To further understand the functions of these genes, we assessed the interactions between 24 metastasis-related genes and 13 TFs (, ). GO analysis was then carried out to determine the functional enrichment of the nodes in this co-expression network (). Several biological processes (BP) and cellular component (CC) pathways were found to be significantly enriched. According to BP, the nodes were mainly enriched in “catecholamine metabolic process”, “lymphocyte proliferation”, “mononuclear cell proliferation”, “lymphocyte differentiation” and “leukocyte proliferation”, while for CC they were enriched in, “integral component of synaptic membrane”, “intrinsic component of synaptic membrane”, and “glutamatergic synapse”.
Table 1 Transcription Factors Associated with 24 Intersecting mRNAs
GSVA Analysis of Seven Prognosis-Related lncRNAs
We performed GSVA to figure out the dynamics of biological processes and pathways for Hallmark gene sets based on seven prognosis-related lncRNAs. As showed, “G2M_CHECKPOINT”, “DNA_REPAIR” and “PI3K_AKT_MTOR_SIGNALING” were remarkably activated in the high AL512422.1 group, while the low AL512422.1 group was enriched in “WNT_BETA_CATENIN_SIGNALING” and “KRAS_SIGNALING_DN”. Upregulation of AL008718.3 activated “WNT_BETA_CATENIN_SIGNALING” and “TGF_BETA_SIGNALING”, but downregulation of AL008718.3 led to “HEDGEHOG_SIGNALING” and “PI3K_AKT_MTOR_SIGNALING”. In addition, AC006033.2 high level was significantly enriched for “HYPOXIA”, “APOPTOSIS” and “NOTCH_SIGNALING”, whereas low level triggered “DNA_REPAIR” and “G2M_CHECKPOINT” process. Notably, “GLYCOLYSIS” and “NOTCH_SIGNALING” were all enriched in AL357507.1 and AL360182.2 low groups. For the C5orf66-AS1 low group, “TGF_BETA_SIGNALING”, “HYPOXIA” and “NOTCH_SIGNALING” were enriched. The C5orf66-AS1 high group is related to “WNT_BETA_CATENIN_SIGNALING”. At last, we observed enrichment of “G2M_CHECKPOINT”, “DNA_REPAIR” and “PI3K_AKT_MTOR_SIGNALING” in the CEBPA-DT high group and “WNT_BETA_CATENIN_SIGNALING” in the low group. The top high-frequency enriched pathways were “NOTCH_SIGNALING”, “HYPOXIA”, “WNT_BETA_CATENIN_SIGNALING” and “HEDGEHOG_SIGNALING”.
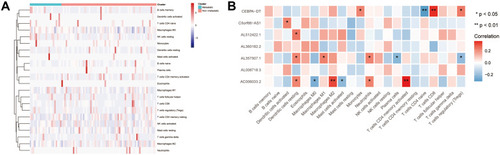
Correlation Between Prognosis-Related lncRNAs and Infiltrating Immunocyte Fractions
To investigate the differences between the primary and metastatic OS, 22 infiltrating immunocyte fractions were compared. As shown in , the difference between the two groups was insignificant. Then, the relationship between seven prognosis-related lncRNAs and 22 infiltrating immune cells was analyzed (). AC006033.2 was found to have the strongest relationship with resting dendritic cells, macrophages M0, macrophages M2, activated mast cells, activated neutrophils, and memory T cells CD4. AL357507.1 had a close relationship with dendritic cells resting, macrophages M1, neutrophils and plasma cells. CEBPA-DT was associated with monocyte, T cells CD4 naïve, T cells CD8 and T cells regulatory (Tregs). C5orf66-AS1 and AL512422.1 were only related to dendritic cells activated and dendritic cells resting, respectively. On the contrary, for AL360182.2 and AL008718.3, there was no association with immune cells.
mRNA Expression of Seven lncRNAs in OS Cell Lines
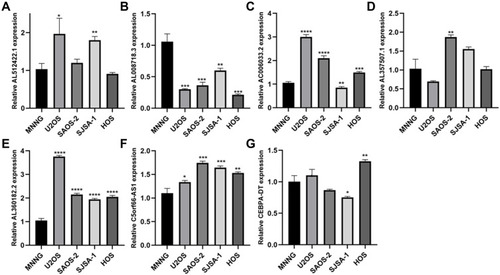
The mRNA level of seven lncRNAs (AL512422.1, AL008718.3, C5orf66-AS1, AL360182.2, CEBPA-DT, AC006033.2, and AL357507.1) in five OS cell lines were examined. MNNG cell line was found to have a high OS metastasis rate, while the other four had a low OS metastasis rate (). AL512422.1 was found to be highly expressed in the U2OS and SJSA-1 cell lines than MNNG (). AL360182.2 and C5orf66-AS1 mRNA levels were found to be higher in four weakly metastatic cell lines (). However, AL008718.3 mRNA level was significantly down-regulated in USO2, SAOS-2, SJSA-1, and HOS cell lines (). In addition, the mRNA level of AL357507.1 was only up-regulated in the SAOS-2 cell line (). Interestingly, even though they were all weak metastatic cell lines, the expression pattern differed. The mRNA level of AC006033.2 was higher in U2OS, SAOS-2, and HOS cell lines, and lower in SJSA-1 cell lines (). In the case of CEBPA-DT, the mRNA levels were higher in the HOS cell line and lower in the SJSA-1 cell line ().
Discussion
OS is an aggressive malignancy with poor outcomes, particularly in patients with metastases.Citation29 The development of advanced treatments has resulted in significant improvement in the overall survival of patients with OS. Surgery is still the cornerstone of primary tumor treatment, whereas adjuvant chemotherapy is the most significant predictor of prognosis in patients with metastases.Citation30,Citation31 Chemotherapies, such as high-dose methotrexate, doxorubicin, cisplatin, and ifosfamide/etoposide are the most effective agents. However, the incidence of metastasis remains high in a significant proportion of OS patients.Citation31 Therefore, to predict or inhibit the metastatic process, it is essential to identify targets to provide potential diagnostic and therapeutic strategies.Citation32
Accumulating evidence suggests that lncRNAs are closely associated with tumorigenesis and progression. lncRNAs play an essential role in a variety of biological processes implicated in carcinogenesis, including cell proliferation, migration, and tumor metastasis.Citation33–Citation36 Identifying an effective lncRNA biomarker may aid in the prevention of metastasis. For example, mtlncRNA LIPCAR is a novel biomarker for cardiac remodeling that can predict the survival rate of heart failure patients.Citation37 Immunoreactive mice showed that targeted therapy of ASncmtRNAs could prevent lung metastases following melanoma resection.Citation38 The lentiviral construct targeting ASncmtRNAs could inhibit B16F10 primary tumor proliferation.Citation39 In vivo studies in isogenic mouse renal cancer models revealed that ASncmtRNAs-targeting ASOs could completely reverse tumor growth.Citation40 In summary, lncRNA targeted therapy may be a potential therapeutic approach for improving prognosis.
In this study, we established a new risk-score model based on prognosis-related lncRNA generated from a series of bioinformatics analyses to predict the metastasis of OS patients. Based on their median risk score, all patients were classified as high or low-risk groups. In comparison to the low-risk group, patients in the high group had a poor survival rate. By calculating the AUC value, this model exhibited a strong prediction capacity. The co-expression network revealed an association between TF and 24 prognosis-related mRNAs, with the seven lncRNAs having the closest relationship. Zhong et al reported that HSF2 was involved in the growth and metastasis of lung cancer.Citation41 It is important to note that the most common site of OS metastasis in the lungs and lung metastases are associated with poor prognosis.Citation30 NRF2 is a transcription factor that modulates the level of reactive oxygen species (ROS), detoxifying agents, and antioxidants in the body. The increase in NRF2 in the nuclei is closely related to bone metastases in OS patients.Citation42 Furthermore, Cindy et al revealed that catecholamines and their receptors could be potential molecular markers for OS progression.Citation43 The catecholamine metabolic process and catechol-containing compound metabolic process were reported as a consequence of 24 prognosis-related mRNA enrichment.
Several overlapping pathways were discovered among the seven lncRNAs enrichment findings, confirming the critical biological function of the seven lncRNAs. The terms “NOTCH_SIGNALING”, “HYPOXIA”, “WNT_BETA_CATENIN_SIGNALING”, and “HEDGEHOG_SIGNALING” appeared more often. Several studies have shown that the Notch pathway regulates OS metastasis and may be useful for therapeutic targeting in the near future.Citation44,Citation45 Fang et al reported that the Notch pathway could be pharmacologically inhibited, which was a huge step in targeted therapeutic strategies.Citation46 CNOT1, in collaboration with LMNA, may exacerbate OS tumorigenesis through the Hedgehog signaling pathway.Citation47 In addition, the hypoxia-HIF-1α-CXCR4 pathway plays a critical role during the migration of human OS cells.Citation48 Furthermore, NRP2 was shown to be overexpressed in OS cell lines; depleting NRP2 through downregulation of the active Wnt-signaling pathway would significantly reduce the tumor burden and metastasis rate in OS cell lines.Citation49 Additionally, we found that three of the seven lncRNAs were associated with resting dendritic cells. According to Jones et al, IL23 expression and monocyte-derived dendritic cells have both activating and suppressive effects on cytotoxic T cells, as well as direct effects on osteoblasts or associated osteoclasts.Citation50 It is worth mentioning that under hypoxic conditions, dendritic cells secrete significant amounts of osteopontin, which contributes to tumor metastasis.Citation51
There were some limitations in the present study. One limitation is the relatively small sample size, even though recruitment is still ongoing. Another limitation is that no additional analysis stratified by age was conducted to account for metastasis. In conclusion, we established a novel model with excellent reliability and accuracy for predicting the metastasis of OS patients.
Abbreviation
AUC, area under receiver operating characteristic curve; BP, biological processes; CC, cellular components; EMT, epithelial–mesenchymal transition; FDR, false discovery rate; GSEA, Gene set enrichment analysis; GSVA, Gene Set Variation Analysis; KM, Kaplan–Meier; LASSO, least absolute shrinkage and selection operator; lncRNAs, long non-coding RNAs; MsigDB, Molecular Signatures Database; OS, osteosarcoma; ROS, reactive oxygen species; TF, transcription factor; Tregs, T cells regulatory.
Data Sharing Statement
Publicly available datasets from the TARGET datasets (https://ocg.cancer.gov/programs/target) were analyzed in this study. The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics Statement
The study was approved by the Ethical Committee of the Affiliated Hospital of Qingdao University and experiments were performed in accordance with the Ethical Committee’s guidelines and regulations. The study complies with guidelines by the Declaration of Helsinki.
Author Contributions
Zhang H, Chen GH and Lyu CY designed the study, acquired the data, and wrote the manuscript. Xu Y reviewed the manuscript. Lyu XJ and Rong C contributed to the Statistical analysis and graphing. Wang YZ and Lyu CY supervised the study. Zhang H and Chen GH contribute equally to this paper. All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
Authors declare that they have no conflict of interest.
References
- Dorfman HD, Czerniak BJ. Bone cancers. Cancer. 1995;75(S1):203–210.8000997
- Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115(7):1531–1543. doi:10.1002/cncr.2412119197972
- Unni KK, Inwards CY. Dahlin’s Bone Tumors: General Aspects and Data on 10,165 Cases. Lippincott Williams & Wilkins; 2010.
- Larsson SE, Lorentzon RJ. The incidence of malignant primary bone tumours in relation to age, sex and site: a study of osteogenic sarcoma, chondrosarcoma and Ewing’s sarcoma diagnosed in Sweden from 1958 to 1968. J Bone Joint Surg Br. 1974;56(3):534–540. doi:10.1302/0301-620X.56B3.534
- Polednak AP. Primary bone cancer incidence in black and white residents of New York State. Cancer. 1985;55(12):2883–2888. doi:10.1002/1097-0142(19850615)55:12<2883::AID-CNCR2820551231>3.0.CO;2-Q3857961
- Ji J, Hemminki K. Familial risk for histology-specific bone cancers: an updated study in Sweden. Eur J Cancer. 2006;42(14):2343–2349. doi:10.1016/j.ejca.2005.11.04316859907
- Bacci G, Longhi A, Fagioli F, et al. Adjuvant and neoadjuvant chemotherapy for osteosarcoma of the extremities: 27 year experience at Rizzoli Institute, Italy. Cancer. 2005;41(18):2836–2845.
- Link MP, Goorin AM, Miser AW, et al. The effect of adjuvant chemotherapy on relapse-free survival in patients with osteosarcoma of the extremity. N Engl J Med. 1986;314(25):1600–1606.3520317
- Rosen G, Caparros B, Huvos AG, et al. Preoperative chemotherapy for osteogenic sarcoma: selection of postoperative adjuvant chemotherapy based on the response of the primary tumor to preoperative chemotherapy. Cancer. 1982;49(6):1221–1230. doi:10.1002/1097-0142(19820315)49:6<1221::AID-CNCR2820490625>3.0.CO;2-E6174200
- Dong J, Liu Y, Liao W, et al. miRNA-223 is a potential diagnostic and prognostic marker for osteosarcoma. J Bone Oncol. 2016;5(2):74–79. doi:10.1016/j.jbo.2016.05.00127335775
- Wang M, Xie R, Si H, et al. Integrated bioinformatics analysis of miRNA expression in osteosarcoma. Artif Cells Nanomed Biotechnol. 2017;45(5):936–943. doi:10.1080/21691401.2016.119645627315542
- Di Fiore R, Drago‐Ferrante R, Pentimalli F, et al. Let‐7d miRNA shows both antioncogenic and oncogenic functions in osteosarcoma‐derived 3AB‐OS cancer stem cells. J Cell Physiol. 2016;231(8):1832–1841. doi:10.1002/jcp.2529126679758
- Schwartz CL, Gorlick R, Teot L, et al. Multiple drug resistance in osteogenic sarcoma: INT0133 from the Children’s Oncology Group. J Clin Oncol. 2007;25(15):2057–2062. doi:10.1200/JCO.2006.07.777617513810
- Gill J, Ahluwalia MK, Geller D, et al. New targets and approaches in osteosarcoma. Pharmacol Ther. 2013;137(1):89–99. doi:10.1016/j.pharmthera.2012.09.00322983152
- Shi J, Dong B, Cao J, et al. Long non-coding RNA in glioma: signaling pathways. Oncotarget. 2017;8(16):27582. doi:10.18632/oncotarget.1517528187439
- Idogawa M, Ohashi T, Sasaki Y, et al. Long non‐coding RNA NEAT1 is a transcriptional target of p53 and modulates p53‐induced transactivation and tumor‐suppressor function. Int J Cancer. 2017;140(12):2785–2791. doi:10.1002/ijc.3068928295289
- Liang Y, Zhang D, Zheng T, et al. lncRNA-SOX2OT promotes hepatocellular carcinoma invasion and metastasis through miR-122-5p-mediated activation of PKM2. Oncogenesis. 2020;9(5):54. doi:10.1038/s41389-020-0242-z32467565
- Gooding AJ, Zhang B, Jahanbani FK, et al. The lncRNA BORG Drives Breast Cancer Metastasis and Disease Recurrence. Sci Rep. 2017;7(1):12698. doi:10.1038/s41598-017-12716-628983112
- Wang Z, Yang B, Zhang M, et al. lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer Cell. 2018;33(4):548.
- Kitamura T, Qian BZ, Soong D, et al. CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J Exp Med. 2015;212(7):1043–1059. doi:10.1084/jem.2014183626056232
- Li C, Wang S, Xing Z, et al. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat Cell Biol. 2017;19(2):106–119. doi:10.1038/ncb346428114269
- Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin β 4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi:10.1038/sj.onc.120692812970751
- Shen L, Chen L, Wang Y, et al. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol. 2015;121(1):101–108. doi:10.1007/s11060-014-1613-025217850
- Ying L, Chen Q, Wang Y, et al. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8(9):2289–2294. doi:10.1039/c2mb25070e22722759
- Fan Y, Shen B, Tan M, et al. TGF-β–induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20(6):1531–1541. doi:10.1158/1078-0432.CCR-13-145524449823
- Kim K, Jutooru I, Chadalapaka G, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32(13):1616–1625. doi:10.1038/onc.2012.19322614017
- Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48(3):R45–R53. doi:10.1530/JME-12-000822393162
- Peng W, Si S, Zhang Q, et al. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Mol Endocrinol. 2015;34(1):1–10.
- Hou CH, Lin FL, Hou SM, et al. Cyr61 promotes epithelial-mesenchymal transition and tumor metastasis of osteosarcoma by Raf-1/MEK/ERK/Elk-1/TWIST-1 signaling pathway. Mol Cancer. 2014;13(1):1–13. doi:10.1186/1476-4598-13-23624387052
- Geller DS, Gorlick R. Osteosarcoma: a review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol. 2010;8(10):705–718.21317869
- Ferguson WS, Goorin AM. Current treatment of osteosarcoma. Cancer Invest. 2001;19(3):292–315. doi:10.1081/CNV-10010255711338887
- He M, Wang G, Jiang L, et al. miR-486 suppresses the development of osteosarcoma by regulating PKC-δ pathway. Int J Oncol. 2017;50(5):1590–1600. doi:10.3892/ijo.2017.392828339053
- Li G, Zhang H, Wan X, et al. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. Biomed Res Int. 2014;2014:780521.24757675
- Maass PG, Luft FC, Bähring S. Long non-coding RNA in health and disease. J Mol Med (Berl). 2014;92(4):337–346. doi:10.1007/s00109-014-1131-824531795
- Tripathi V, Shen Z, Chakraborty A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9(3):e1003368. doi:10.1371/journal.pgen.100336823555285
- Zhang A, Xu M, Mo YY. Role of the lncRNA-p53 regulatory network in cancer. J Mol Cell Biol. 2014;Jun(3):181–191. doi:10.1093/jmcb/mju013
- Kumarswamy R, Bauters C, Volkmann I, et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014;114(10):1569–1575. doi:10.1161/CIRCRESAHA.114.30391524663402
- Lobos-González L, Silva V, Araya M, et al. Targeting antisense mitochondrial ncRNAs inhibits murine melanoma tumor growth and metastasis through reduction in survival and invasion factors. Oncotarget. 2016;7(36):58331. doi:10.18632/oncotarget.1111027507060
- Varas‐Godoy M, Lladser A, Farfan N, et al. In vivo knockdown of antisense non‐coding mitochondrial RNA s by a lentiviral‐encoded sh RNA inhibits melanoma tumor growth and lung colonization. Pigment Cell Melanoma Res. 2018;31(1):64–72. doi:10.1111/pcmr.1261528707763
- Borgna V, Villegas J, Burzio VA, et al. Mitochondrial ASncmtRNA-1 and ASncmtRNA-2 as potent targets to inhibit tumor growth and metastasis in the RenCa murine renal adenocarcinoma model. Oncotarget. 2017;8(27):43692. doi:10.18632/oncotarget.1846028620146
- Zhong YH, Cheng HZ, Peng H, et al. Heat shock factor 2 is associated with the occurrence of lung cancer by enhancing the expression of heat shock proteins. Oncol Lett. 2016;12(6):5106–5112. doi:10.3892/ol.2016.536828101237
- Zhang J, Wang X, Wu W, et al. Expression of the Nrf2 and Keap1 proteins and their clinical significance in osteosarcoma. Biochem Biophys Res Commun. 2016;473(1):42–46. doi:10.1016/j.bbrc.2016.03.04726987716
- Bandala C, Ávila-luna A, Gómez-López M, et al. Catecholamine levels and gene expression of their receptors in tissues of adults with osteosarcoma. Arch Physiol Biochem. 2019;10:1–7.
- Tanaka M, Setoguchi T, Hirotsu M, et al. Inhibition of Notch pathway prevents osteosarcoma growth by cell cycle regulation. Br J Cancer. 2009;100(12):1957–1965. doi:10.1038/sj.bjc.660506019455146
- Dai G, Liu G, Zheng D, et al. Inhibition of the Notch signaling pathway attenuates progression of cell motility, metastasis, and epithelial-to-mesenchymal transition-like phenomena induced by low concentrations of cisplatin in osteosarcoma. Eur J Pharmacol. 2021;15(899):174058. doi:10.1016/j.ejphar.2021.174058
- Fang L, Li B, Yu D, et al. Analysis of changes in the expression of Notch1 and HES1 and the prognosis of osteosarcoma patients following surgery. Oncol Lett. 2020;20(4):1. doi:10.3892/ol.2020.1215632774475
- Cheng D, Li J, Li S, et al. CNOT 1 cooperates with LMNA to aggravate osteosarcoma tumorigenesis through the Hedgehog signaling pathway. Mol Oncol. 2017;11(4):388–404. doi:10.1002/1878-0261.1204328188704
- Guo M, Cai C, Zhao G, et al. Hypoxia promotes migration and induces CXCR4 expression via HIF-1α activation in human osteosarcoma. PLoS One. 2014;9(3):e90518. doi:10.1371/journal.pone.009051824618817
- Ji T, Guo Y, Kim K, et al. Neuropilin-2 expression is inhibited by secreted Wnt antagonists and its down-regulation is associated with reduced tumor growth and metastasis in osteosarcoma. Mol Cancer. 2015;14(1):1–14. doi:10.1186/s12943-015-0359-425560632
- Jones KB. Dendritic Cells Drive Osteosarcomagenesis through Newly Identified Oncogene and Tumor Suppressor. Cancer Discov. 2019;9(11):1484–1486. doi:10.1158/2159-8290.CD-19-099431676561
- Yang M, Ma C, Liu S, et al. Hypoxia skews dendritic cells to a T helper type 2-stimulating phenotype and promotes tumour cell migration by dendritic cell-derived osteopontin. Immunology. 2009;128(1pt2):e237–e249. doi:10.1111/j.1365-2567.2008.02954.x19740309