Abstract
The Philadelphia chromosome (Ph)-negative myeloproliferative neoplasms (MPNs), including polycythemia vera, essential thrombocythemia, and primary myelofibrosis, are a group of clonal hematopoietic stem cell disorders with overlapping clinical and cytogenetic features and a variable tendency to evolve into acute leukemia. These diseases not only share overlapping chromosomal abnormalities but also a number of acquired somatic mutations. Recently, mutations in a putative tumor suppressor gene, ten-eleven translocation 2 (TET2) on chromosome 4q24 have been identified in 12% of patients with MPN. Additionally 4q24 chromosomal rearrangements in MPN, including TET2 deletions, have also been observed using conventional cytogenetics. The goal of this study was to investigate the frequency of genomic TET2 rearrangements in MPN using fluorescence in situ hybridization as a more sensitive method for screening and identifying genomic deletions. Among 146 MPN patients, we identified two patients (1.4%) who showed a common 4q24 deletion, including TET2. Our observations also indicated that the frequency of TET2 deletion is increased in patients with an abnormal karyotype (5%).
Introduction
The Philadelphia chromosome negative (Ph-) myeloproliferative neoplasms (MPNs) are a group of clonal hematopoietic stem cell disorders with overlapping clinical and cytogenetic features and a variable tendency to evolve into acute leukemia. The Ph- MPNs include polycythemia vera, essential thrombocythemia, and primary myelofibrosis. The current unifying concept of cytogenetic instability of Ph- MPN is a loss or gain of chromosomal regions. The most frequent recurrent chromosomal abnormalities occurring in the Ph- MPNs include trisomy 9/+9p, deletion (20) (q11q13), deletion of 13q, and trisomy of chromosomes 8 and 9. The prognostic significance of the abnormalities in MPNs remains uncertain. However, recent attempts have been made to classify cytogenetic findings in primary myelofibrosis patients as being associated with favorable [isolated deletion (20q) and 13q, trisomy 9 as well as normal karyotype] outcomes and unfavorable outcomes (all other chromosome abnormalities). The number of patients remains too small and the time of follow-up too limited for a definitive validation of the outcomes associated with chromosomal risk profiles.Citation1 Nevertheless, it appears that the presence of cytogenetic abnormalities in primary myelofibrosis is associated with shorter survival, but their independent contribution to patient prognosis remains unknown.Citation2
Besides cytogenetic abnormalities, these diseases also share acquired somatic mutation of JAK2 in exon 14, resulting in a valine to phenylalanine substitution in codon 617, JAK2V617F, present in over 90% of patients with polycythemia vera, 60% of patients with essential thrombocythemia, and 50% of patients with primary myelofibrosis.Citation3–Citation6 Although most MPNs carry mutation on one of two alleles, 2.8% of MPNs acquire JAK2V617F on two JAK2 alleles,Citation7 and 3% of JAK2V617F-negative patients have mutations in exon 12 of JAK2.Citation8 Coexistence of these two mutations have also been described.Citation9 MPL mutations leading to MPLW515L occur in 5%–10% of patients with primary myelofibrosis and in 2%–5% of patients with essential thrombocythemia who are negative for JAK2V617F.Citation10,Citation11 Other mutations described in MPN patients include mutations of the CBL and ASXL1 genes.Citation12,Citation13
Recently, somatic mutations and deletions of the ten-eleven translocation 2, or TET2, a putative tumor suppressor gene located on chromosome 4, band q24, have been described in 12% of patients with MPN.Citation14 TET2 mutations may predate the JAK2V617F mutation, may occur after the acquisition of JAK2V617F, or may occur simultaneously in two different clones.Citation14,Citation15 Following the original description, TET2 nonsense and splice mutations, deletions, and out-of-frame insertions were identified in other myeloid malignancies, including myelodysplastic syndrome (19%–26%), de novo acute myeloid leukemia (12%–20%), therapy-related acute myeloid leukemia (24%–43%), chronic myelomonocytic leukemia (20%–51%), and systemic mastocytosis (29%).Citation14,Citation16–Citation24 Although the current prognostic value of TET2 mutations in MPNs remains unknown, TET2 mutations were reported in 6/14 patients (43%) with MPN who had transformed to acute myeloid leukemia.Citation25
Deletions of the 4q24 region, including the TET2 gene, have been identified by conventional cytogenetics. In 2005, Viguie et al reported a common deletion of 4q24 in four patients with acute myeloid leukemia/myelodysplastic syndrome.Citation26 Additional cytogenetic and high-resolution single-nucleotide polymorphism karyotyping revealed that 20/886 (2%) patients with myeloid disorders exhibited either partial or complete deletion of TET2.Citation14,Citation18–Citation20 In at least four patients with acute myeloid leukemia/myelodysplastic syndrome, the remaining TET2 copy harbored a somatic mutation.Citation19 More recently, Hussein compared cytogenetic findings with TET2 status and concluded that unmutated TET2 MPN patients were not cytogenetically different from those MPN patients who harbor TET2 mutations, but there was no information provided regarding deletions of 4q24/TET2.Citation27
Materials and methods
Patients
A total of 146 patient samples (48 were obtained from the Myeloproliferative Disorders-Research Consortium Tissue Bank) were entered in this study, including myelofibrosis (n = 52), polycythemia vera (n = 47), essential thrombocythemia (n = 21), myeloproliferative neoplasm (unclassified, MPNu, n = 20), and MPNu/myelodysplastic syndrome (n = 6). After patient informed consent was obtained, peripheral blood and bone marrow specimens were collected according to the Institutional Review Board guidelines of the Mount Sinai School of Medicine.
Bacterial artificial chromosome clones
Three bacterial artificial chromosome (BAC) clones, RP11-912N16 (137 kb), RP11-16G16 (175 kb), and RP11-45L9 (162 kb) obtained from BACPAC Resources, Oakland, CA, were grown on chloramphenicol agar plates. The BAC clone RP11-16G16 contains the commonly deleted region on 4q24, including TET2. To determine the extent of the deletion or possible rearrangements of the 4q24 region, two additional BAC clones, RP11-912N16 and RP11-45L9, containing DNA segments centromeric and telomeric to this commonly deleted region, respectively, were also used. Selected positive colonies were harvested after overnight growth using the Qiagen Large-Construct kit (Qiagen, Valenica, CA). DNA labeling was achieved using the CGH Nick Translation Kit (Abbott Molecular, Des Plaines, IL) and was fluorescently labeled with spectrum green (RP11-912N16), spectrum aqua (RP11-16G16), and spectrum red (RP11-45L9) according to the Abbott Molecular CGH protocol (Abbott Molecular). Validation fluorescence in situ hybridization (FISH) studies for all three BAC clones were performed on normal controls by evaluating 2000 interphase nuclei. The cutoff percentage for these probes was 3.5% and was calculated by using the mean ± three standard deviations. All three BAC probes mapped to the 4q24 region in 40 examined metaphase cells, demonstrating the 100% specificity of these probes. In interphase nuclei of normal cells, the expected normal probe signals appear as two tricolor (green, red, aqua) fusions.
Cytogenetic and FISH analysis
FISH analysis was performed as previously described with pepsin modification treatment (100 uL 10% pepsin and 2 mL 1% HCl) for five minutes.Citation28 Codenaturation of cells and FISH probes were carried out on a Thermobrite (Abbott Molecular) for three minutes at 73°C and hybridized at 37°C overnight. A total of 200 nuclei were scored in each of the 146 specimens. Metaphase cells were obtained using standard cytogenetic methods. Chromosomal abnormalities were described according to the International System of Human Cytogenetic Nomenclature (2009).Citation29
Results
We investigated 146 patients with Ph- MPN to determine the frequency of TET2 genomic rearrangements. The summary of FISH studies, using the three TET2 BAC FISH probes as well as the cytogenetic results, are provided in . Chromosomal analyses were available for 83/146 (57%) patients; 44 (53%) patients had a normal karyotype and 39 (47%) had an abnormal karyotype. The frequency of chromosomal abnormalities among the 39 patients with an abnormal karyotype were: deletion of the long arms of chromosome 20 in 11/39 (29%), followed by +9/+i(9p) in 9/39 (24%), trisomy of chromosome 8 in 7/39 (18%), and trisomy of the long arms of chromosome 1 in 6/39 (16%). Abnormalities of 4q24 (deletions, translocations, or duplications) were not detected in any of the 39 patients with an abnormal karyotype by conventional cytogenetics. Interphase FISH analysis using these probes revealed deletion of TET2 in 2/83 patients (2% of total, and 5% among cytogenetically abnormal).
Table 1 Interphase FISH detection of TET2 deletion
Patient 34 with primary myelofibrosis was JAK2V617F+ and had a complex karyotype, but 4q24 rearrangement was not identified (see ). Interphase FISH with TET2 revealed a deletion of all three BAC probes in 5% of interphase nuclei. The follow-up peripheral blood specimen eight months later revealed 30% of interphase cells with a TET2 deletion, as well as a clone with a cytogenetically identified deletion of the 4q24 region (see ). Patient 8, with primary myelofibrosis, was JAK2V617F+, had a normal karyotype (see ), and was disomic for the following loci: 7p11.1–q11.1, 8p11.1–q11.1, 9p11.1–q11.1, 1q12, 1q21, 1p36, 1q25, 5p15.2, 5q31, 7q31, 9p21 13q14, and 20q12 using interphase FISH. These loci are most frequently rearranged in MPN. In contrast, 90% of interphase cells revealed a deletion of only the RP11-16G16 BAC probe, and not RP11-912N16 and RP11-45L9, indicating a small interstitial TET2 deletion of at least 175 kb localized between the centromeric and telomeric BAC FISH probes (see ).
Table 2 Summary of results from two patients with TET2 deletions
Figure 1 Interphase nuclei representing TET2 FISH signal patterns. A) Normal nuclei with two triple color fusion signals. B) Deletion of the 175 kb RP11-16G16 BAC probe (aqua). C) Deletion of all three bacterial artificial chromosome probes; note only one triple color fusion signal.
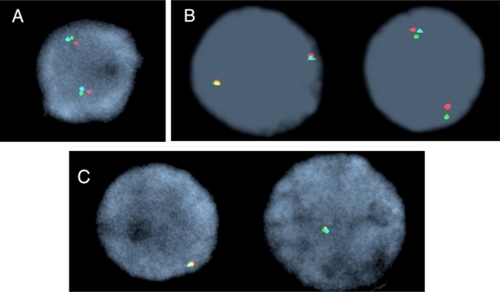
Based on these initial studies, we screened an additional 63 patients for TET2 genomic rearrangements, as shown in , and none were identified.
also shows JAK2 status in 42 patients and MPLW515L mutational status in 16 patients. Of the three patients who had TET2 deletion, one was MPLW515L mutation-negative, and the MPL mutational status of the remaining two patients was not available.
Among 22,500 cytogenetically examined patients at our institution we identified only three patients with 4q24 chromosomal rearrangements, two with myelodysplastic syndrome and one with primary myelofibrosis (#34), indicating their rare occurrence. Patient 34 was identified among 512 patients with MPN (polycythemia vera = 361, myelofibrosis = 151). We therefore used interphase FISH as a more sensitive method to detect 4q24 chromosomal rearrangements and identified an additional patient of the 146 tested patients with TET2 deletion, confirming the published frequency of 1%–2% in MPNs.Citation14,Citation18 However, among patients with available conventional cytogenetic results, the frequency was 2% overall (2/83), or 5% (2/39) among the cytogenetically abnormal patients. Two patients had primary myelofibrosis, while none of the 47 patients with polycythemia vera nor 21 patients with essential thrombocythemia had TET2 deletions.
The size of the deletions varied. In one patient (#8) with a normal karyotype, a deletion of approximately 175 kb in 90% of interphase nuclei containing the commonly deleted region of 4q24 (the other two BACs outside the region remained intact) was identified. This observation suggests a submicroscopic (cryptic) deletion beyond the resolution of conventional cytogenetic analysis and confirms the importance of using a higher resolution methodology for identifying TET2 deletion. Moreover, metaphase FISH analysis confirmed a submicroscopic 4q24 deletion that was not detectable by conventional cytogenetics. The other patient (#34) had a deletion of all three BACs, resulting in a loss of at least 1.6 Mb. Interestingly, patient 34 had initially 5% of interphase cells with TET2 deletion that was not detectable by conventional cytogenetics. When 30% of his peripheral blood cells showed loss of TET2, a new complex cytogenetic clone emerged showing der(4)del(4)(p14)del(q24), resulting in a complete 4q24 deletion.
Heterozygous TET2 mutations have been rarely identified in patients with chronic myelogenous leukemia but were detected in those patients who progress to blast crisis or accelerated phase.Citation30 Interpretation of these observations suggested that TET2 mutations represent secondary lesions which contribute to the progression of the disease.Citation30 We identified one patient with chronic myelogenous leukemia, not included in this study, who had complex chromosomal aberrations consistent with blast crisis of chronic myelogenous leukemia. FISH analysis revealed a complete deletion of all three BAC probes in 17% of interphase nuclei.
In summary, TET2 deletions are not a frequent genomic event in MPN. Their deletions are present in about 2% of MPN patients and, in those with increased karyotype instability, they may occur in about 5% of patients. These observations may add supporting evidence that TET2 mutations, as well as deletions, may contribute to leukemic evolution in patients with MPN that transform into leukemia.
Acknowledgements
This work was supported in part by the P01 CA 108671 MPD-RC grant from the National Institutes of Health.
Disclosure
The authors report no conflicts of interest in this work.
References
- HusseinKvan DykeDLTefferiAConventional cytogenetics in myelofibrosis: Literature review and discussionEur J Haematol20098232933819141119
- CervantesFDupriezBPereiraANew prognostic scoring system for primary myelofibrosis based on a study of the International Working Group for Myelofibrosis Research and TreatmentBlood20091132895290118988864
- JamesCUgoVLe CouédicJPA unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia veraNature20054341144114815793561
- KralovicsRPassamontiFBuserASA gain-of-function mutation of JAK2 in myeloproliferative disordersN Engl J Med20053521779179015858187
- LevineRLWadleighMCoolsJActivating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosisCancer Cell2005738739715837627
- BaxterEJScottLMCampbellPJAcquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disordersLancet20053651054106115781101
- OlcayduDHarutyunyanAJägerRA common JAK2 haplotype confers susceptibility to myeloproliferative neoplasmsNat Genet20094145045419287385
- ScottLMBeerPABenchAJErberWNGreenARPrevalence of JAK2 V617F and exon 12 mutations in polycythaemia veraBr J Haematol200713951151217910642
- LiSKralovicsRde LiberoGTheocharidesAGisslingerHSkodaRCClonal heterogeneity in polycythemia vera patients with JAK2 exon12 and JAK2-V617F mutationsBlood20081113863386618195094
- BeerPACampbellPJScottLMMPL mutations in myeloproliferative disorders: Analysis of the PT-1 cohortBlood200811214114918451306
- ChalignéRJamesCTonettiCEvidence for MPL W515L/K mutations in hematopoietic stem cells in primitive myelofibrosisBlood20071103735374317709604
- GrandFHHidalgo-CurtisCEErnstTFrequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasmsBlood20091136182619219387008
- CarbucciaNMuratiATrouplinVMutations of ASXL1 gene in myeloproliferative neoplasmsLeukemia2009232183218619609284
- DelhommeauFDupontSDella ValleVMutation in TET2 in myeloid cancersN Engl J Med20093602289230119474426
- SchaubFXLooserRLiSClonal analysis of TET2 and JAK2 mutations suggests that TET2 can be a late event in the progression of myeloproliferative neoplasmsBlood20101152003200720061559
- LangemeijerSMKuiperRPBerendsMAcquired mutations in TET2 are common in myelodysplastic syndromesNat Genet20094183884219483684
- TefferiASkodaRVardimanJWMyeloproliferative neoplasms: Contemporary diagnosis using histology and geneticsNat Rev Clin Oncol2009662763719806146
- JankowskaAMSzpurkaHTiuRVLoss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasmsBlood2009113640710
- KosmiderOGelsi-BoyerVCheokMTET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs)Blood20091143285329119666869
- KosmiderOGelsi-BoyerVCiudadMTET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemiaHaematologica2009941676168119797729
- TefferiALimKHLevineRMutation in TET2 in myeloid cancersN Engl J Med20093611117 author reply 1117, 1118.19741235
- TefferiAMolecular drug targets in myeloproliferative neoplasms: Mutant ABL1, JAK2, MPL, KIT, PDGFRA, PDGFRB and FGFR1J Cell Mol Med20091321523719175693
- Saint-MartinCLeroyGDelhommeauFAnalysis of the ten-eleven translocation 2 (TET2) gene in familial myeloproliferative neoplasmsBlood20091141628163219564637
- Abdel-WahabOMullallyAHedvatCGenetic characterization of TET1, TET2, and TET3 alterations in myeloid malignanciesBlood200911414414719420352
- Abdel-WahabOManshouriTPatelJGenetic analysis of transforming events that convert chronic myeloproliferative neoplasms to leukemiasCancer Res20107044745220068184
- ViguiéFAbouraABouscaryDCommon 4q24 deletion in four cases of hematopoietic malignancy: Early stem cell involvement?Leukemia2005191411141515920487
- HusseinKAbdel-WahabOLashoTLCytogenetic correlates of TET2 mutations in 199 patients with myeloproliferative neoplasmsAm J Hematol201085818319957346
- NajfeldVMontellaLScaliseAFruchtmanSExploring polycythaemia vera with fluorescence in situ hybridization: Additional cryptic 9p is the most frequent abnormality detectedBr J Haematol200211955856612406101
- ShafferLGSlovakMLCampbellLJISCN (2009): International System of Human Cytogenetic NomenclatureBaselS Karger AG2009
- MakishimaHJankowskaAMCazzolliHCbl and TET2 mutations are present in refractory Ph+ disorders including accelerated and blast crisis CML and ALLBlood20091142173