Abstract
Purpose
There is currently no measure to predict a treatability of long-acting β-2 agonist (LABA) or long-acting muscarinic antagonist (LAMA) in patients with chronic obstructive pulmonary disease (COPD). We aimed to build prediction models for the treatment response to these bronchodilators, in order to determine the most responsive medication for patients with COPD.
Methods
We performed a prospective open-label crossover study, in which each long-acting bronchodilator was given in a random order to 65 patients with stable COPD for 4 weeks, with a 4-week washout period in between. We analyzed 14 baseline clinical traits, expression profiles of 31,426 gene transcripts, and damaged-gene scores of 6,464 genes acquired from leukocytes. The gene expression profiles were measured by RNA microarray and the damaged-gene scores were obtained after DNA exome sequencing. Linear regression analyses were performed to build prediction models after using factor and correlation analyses.
Results
Using a prediction model for a LABA, traits found associated with the treatment response were post-bronchodilator forced expiratory volume in 1 second, bronchodilator reversibility (BDR) to salbutamol, expression of three genes (CLN8, PCSK5, and SKP2), and damage scores of four genes (EPG5, FNBP4, SCN10A, and SPTBN5) (R2=0.512, p<0.001). Traits associated with the treatment response to a LAMA were COPD assessment test score, BDR, expression of four genes (C1orf115, KIAA1618, PRKX, and RHOQ) and damage scores of three genes (FBN3, FDFT1, and ZBED6) (R2=0.575, p<0.001). The prediction models consisting only of clinical traits appeared too weak to predict the treatment response, with R2=0.231 for the LABA model and R2=0.121 for the LAMA model.
Conclusion
Adding the expressions of genes and damaged-gene scores to the clinical traits may improve the predictability of treatment response to long-acting bronchodilators.
Introduction
Chronic obstructive pulmonary disease (COPD) is a leading cause of morbidity and mortality worldwide and results in a substantial socioeconomic burden.Citation1 The treatment regimen for a given patient is determined based on the patient’s symptoms and history of exacerbations,Citation2 and long-acting inhaled bronchodilators are the most essential pharmacologic agents. The Global Initiative for Chronic Obstructive Lung Disease (GOLD) guidelines recommend the regular use of a long-acting bronchodilator, either a long-acting β-2 agonist (LABA) or a long-acting muscarinic antagonist (LAMA) for group B COPD patients, that is, symptomatic patients without frequent exacerbations, but there is no clear evidence to recommend one class of long-acting bronchodilators over another.Citation2
The therapeutic efficacy of long-acting bronchodilators in the treatment of stable COPD has been well established.Citation3 They increase forced expiratory volume in 1 second (FEV1) values and improve patient-reported outcomes, such as dyspnea, health-related quality of life, and exercise tolerance in COPD patients.Citation4–Citation9 They also reduce the risks of exacerbations and hospitalizations, compared with placebo.Citation10,Citation11 However, evidence has shown clearly that not all patients respond equally to both.Citation12,Citation13 A considerable number of patients show no response to bronchodilator treatment, and experience adverse effects, such as palpitations and dry mouth, leading to decreased adherence to treatment; thus, identifying potential responders and nonresponders before a bronchodilator treatment is initiated is important.
Previous studies have reported several traits to be related to the variability in response to short-acting β-2 agonists, including age,Citation14 smoking,Citation15 and baseline lung function.Citation16 These findings suggest that individual clinical characteristics are possibly related to the variable responses to bronchodilator treatment. However, contrary to short-acting β-2 agonist treatment, for long-acting bronchodilator treatment, no specific clinical characteristics have been found to be significant predictors of response. Previous reports on the genetic variation between β-2 receptors and muscarinic receptors have suggested that targeted treatment would be beneficial in improving the therapeutic effect,Citation17–Citation19 but single-gene polymorphism was not enough to explain the variation in treatment response in COPD patients.Citation20
If we could predict the treatment response to a particular class of bronchodilator based on patient traits before treatment, we would be better positioned to choose the most responsive medication for individual patients. More importantly, knowing traits associated with treatment response to a LABA or LAMA may expand our understanding of the pathophysiology of COPD and help to identify potential treatment targets. With the idea that genetic traits have a role in treatment response, we aimed to build a prediction model for long-acting bronchodilator treatment response, using patient clinical characteristics, and genetic traits, including gene expression and damaged-gene scores.
Methods
Study design
This was a multicenter, prospective, open-label crossover study aimed at building a prediction model for long-acting bronchodilator treatment response in patients with COPD. Enrolled patients were instructed to discontinue all long-acting bronchodilators for at least 4 weeks, if they were using any. To build the prediction model, the following baseline traits were recorded for each patient: age, body mass index, smoking history, modified Medical Research Council (mMRC) dyspnea scale score, visual analog scale of cough and sputum score, the Korean version of the COPD assessment test (CAT) score, and pre- and post-bronchodilator spirometry (200 μg of salbutamol was administered with a spacer 15 minutes before spirometry testing), diffusing capacity, and lung volume measurements. In addition, blood samples for the expression profiles and damaged-gene scores were obtained from each patient.
Patients were randomly assigned to either a β-2 agonist (150 μg of indacaterol single-dose dry powder inhaler [Onbrez Breezhaler; Norvatis, Basel, Switzerland]) or a muscarinic antagonist (18 μg of tiotropium [Spiriva Handihaler; Boehringer Ingelheim, Ingelheim, Germany]) for the first 4 weeks. Then, after a 4-week washout period in between, the other medication was administered for the final 4 weeks (). Each medication was administered once daily by inhalation in the morning. All patients were given instructions on inhaler device use prior to treatment. They were asked to visit the clinic at weeks 4, 8, and 12 for spirometry testing, and to report respiratory symptoms, concomitant medications, and adverse events if they experienced any. Compliance with inhaler treatment was determined by patient self-report. We categorized the level of compliance into four categories: good compliance =75%–100% of the prescribed inhaler medication was reported used; borderline compliance =50%–75% reported used; poor compliance =25%–50% reported used; and minimal compliance =0%–25%.
Figure 1 Study design.
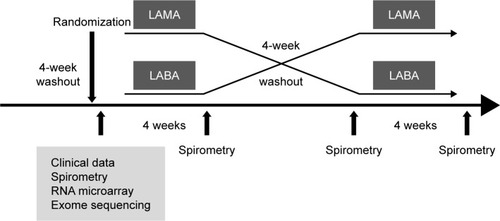
If a patient experienced an acute COPD exacerbation that required a medication modification during the study period, the patient was removed from the study. All respiratory medications were permitted except for other inhaled long-acting bronchodilators, corticosteroids, and macrolide antibiotics. Salbutamol was allowed as a symptom reliever. The objective of the study was to build prediction models for the treatment response to a LABA and LAMA. A significant treatment response was defined as an increase of 100 mL in FEV1.Citation21
Patients
Patients were recruited at five institutions in South Korea. Criteria for inclusion were an age of 45 years or more, a smoking history of at least 10 pack-years, experience of exertional dyspnea for more than a year, and a diagnosis of moderate COPD according to the GOLD criteria (post-bronchodilator FEV1 between 50% and 80% of predicted, in the presence of post-bronchodilator FEV1/forced vital capacity ratio of <0.70).
Exclusion criteria were a COPD exacerbation or respiratory infection in the 4 weeks before screening, a history of asthma, and the presence of bronchiectasis or sequelae of tuberculosis. The study protocol was approved by the Asan Medical Center Institutional Review Board (IRB No 2014-0298). The study was registered with the Clinical Research Information Service (CRIS) before patient enrollment began; CRIS is a not-for-profit online registration system for clinical researchers established by the Korean Centers for Disease Control and Prevention (registration No KCT001092). CRIS is part of the World Health Organization International Clinical Trials Registry Platform. All patients gave written informed consent.
RNA microarray analysis for gene expression
RNA microarray analysis was performed for 31,426 gene transcripts by a specialized company, Macrogen (Seoul, South Korea). The method for RNA preparation and processing is illustrated in the Supplementary material. All data analyses and visualization of differentially expressed genes were conducted using R, version 2.4.1.
DNA exome sequencing
The exome sequencing was also performed by Macrogen (Seoul, South Korea). DNAs extracted from leukocytes were used for whole-exome next-generation sequencing. The method for DNA preparation is illustrated in the Supplementary material. The qPCR Quantification Protocol Guide was used for quantifying the final purified product, and the exome sequencing was performed using the HiSeq™ 2500 platform (Illumina, San Diego, CA, USA).
Damaged-gene score calculation methods
The damaged-gene score represents the overall impact of a nonsynonymous mutation in a gene, with lower scores indicating more damage in the molecular function of the gene. The damaged-gene score was calculated using the sorting intolerant from tolerant (SIFT) algorithm.Citation22 A filtering strategy based on rare (minor allele frequencies in 1,000 genome <0.01) and damaged (SIFT threshold <0.05) variants identified 6,464 genes, and the damaged-gene score was individually computed using the geometric mean of the SIFT score for nonsynonymous variants of the genes. The 1,000 Genome Data is a single-variation catalogue with allele frequencies in 2,504 healthy individuals, and the data have been used as a basis for filtering rare variants in several studies.Citation23 The SIFT score, which indicates the degree of damage of the individual variant, has already been shown in many studies to be indicative of the degree of damage to the gene.Citation24–Citation26 The method of using the damaged-gene score has been used in a previous study on drug responsiveness to damaged genes.Citation27
Analytic validation
Analytic validation was performed for RNA microarray and DNA exome sequencing by Quantitative TaqMan PCR and SNP Type Assay, respectively.
Quantitative TaqMan PCR
Gene expression profiles were validated by Quantitative TaqMan PCR. Validation was performed by Macrogen (Seoul, South Korea). From the genes showing a significant correlation between expression level and treatment response, we arbitrarily selected 28 (Table S1 for the full names of the selected genes), and endogenous controls were glyceraldehyde-3-phosphate dehydrogenase and beta-actin (ACTB). The comparative Ct method was performed for relative quantification. Then we performed Pearson’s correlation analysis to evaluate the accuracy of the RNA microarray results. The higher the correlation between the RNA microarray and quantitative TaqMAN PCR results, the more reliable the microarray results were considered to be. The medians (ranges) of the Pearson’s coefficients were 0.853 (0.792–0.921) and 0.835 (0.638–0.953) for the genes associated with the LABA treatment response and LAMA treatment response, respectively.
SNP type assay
Exome sequencing was validated using SNP Type Assay (Fluidigm, San Francisco, CA, USA). The SNP Type Assay sequencing was performed by a specialized company (DNA Link, Inc., Seoul, South Korea). Mutant alleles detected by whole-exome next-generation sequencing and SNP Type Assay were compared for all genotypes of damaged variants of the four and three genes used in the prediction models for LABA and LAMA treatment, respectively. Genotype was classified as either homozygous or heterozygous in alleles. A total of 33 variants of genes used in the prediction model were found to be completely identical in the two methods.
Statistical analysis
In addition to age, the following 13 clinical traits were chosen for the analysis: body mass index, smoking status (ex-smoker versus current smoker), intensity of smoking (pack-years of cigarettes smoked), CAT score, mMRC dyspnea scale score, pre- and post-bronchodilator FEV1 (%predicted value), bronchodilator reversibility (BDR) to 200 μg of salbutamol (%predicted value), diffusing capacity (% of predicted value), total lung capacity (TLC, % of predicted value), ratio of inspiratory capacity to TLC, residual volume (RV) (% of predicted value), and RV/TLC. We reduced this number from 13 to 6, using a factor analysis, as shown in Table S2. Body mass index, intensity of cigarette smoking, CAT score, post-bronchodilator FEV1, BDR, and RV/TLC were chosen.
The association between gene expression level and treatment response (ie, FEV1 changes after 4 weeks of long-acting bronchodilator treatment) was assessed using Pearson’s correlation analysis, identifying the 28 genes with the highest correlation with improvement in FEV1 values. The 28 genes were further assessed by linear regression models, leaving 7 and 9 genes that showed a statistically significant association with improvement in FEV1 after the LABA and LAMA treatment, respectively. Factor analysis was then done to reduce the number of the genes, with three and four genes selected for the prediction model for LABA and LAMA treatment response, respectively (Figure S1). The damaged-gene scores were also assessed using the same method, ultimately identifying four and three genes for the LABA and LAMA models, respectively (Figure S2).
Multivariate linear regression analysis was performed to build models to predict changes in FEV1 after treatment, using the aforementioned six clinical traits, plus age, level of gene expression, and damaged-gene scores. The prediction performance was assessed using the R2 value.
SPSS, version 21 (IBM Corporation, Armonk, NY, USA), was used for statistical analysis.
Results
Baseline clinical traits
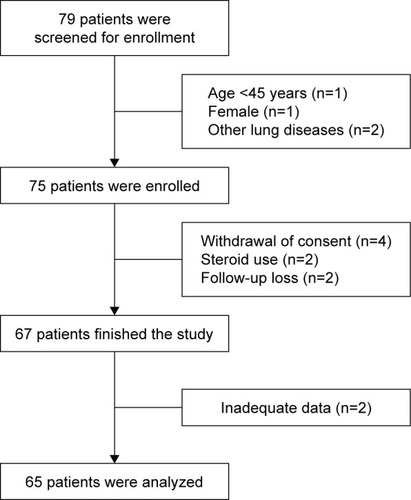
Of the 79 patients screened, 65 were included in the final analysis (). There was only one female patient, and we decided not to include the female patient because of the possibility of confounding. The baseline clinical traits are shown in . The mean age was 67.4 years. Ex-smokers accounted for 69.2% of the patients, whereas the rest were current smokers. The mean number of pack-years was 41.9. More than three-quarters of participants reported grade 1 dyspnea. The mean CAT score was 9.9, and the mean post-bronchodilator FEV1 was 69.8% of the predicted value. Thirty-three patients were randomized to receive the LAMA first, and the rest (32 patients) were given the LABA first.
Table 1 Baseline clinical characteristics of study patients
Response to and compliance with bronchodilator treatment
Of all of the study patients, 34 showed a significant response (FEV1 increase >100 mL) to the LABA and 33 patients to the LAMA. The mean ± SD change in FEV1 after LABA and LAMA treatment was 118±186 and 112±192 mL, respectively. About one-quarter of patients (17/65) showed a significant treatment response to both types of bronchodilators. Conversely, 15 patients had no response to either type.
All 34 LABA responders had good compliance; of the nonresponders, 3 patients (9.7%) were categorized as poor compliance, while the remaining 28 patients (90.3%) had good compliance. Of the 33 LAMA responders, 3 patients (9.1%) had poor compliance, while the remaining 30 patients (90.9%) had good compliance; all 32 nonresponders had good compliance.
Prediction models for long-acting bronchodilator treatment response using clinical traits
The clinical traits were used in the linear regression to build prediction models for the treatment response to the bronchodilators. Among the six models built for each bronchodilator, we chose one that consisted of variables with p-value <0.05. The model of post-bronchodilator FEV1 and BDR could predict FEV1 response after the LABA treatment with statistical significance (R2=0.231, p<0.001) (). Likewise, CAT score and BDR could predict FEV1 response after the LAMA treatment (R2=0.121, p=0.021) ().
Table 2 Linear regression models for predicting FEV1 changes after LABA, using clinical traits
Table 3 Linear regression models for predicting FEV1 changes after LAMA, using clinical traits
Prediction models for long-acting bronchodilator treatment response using gene expression and damaged-gene scores
Three gene expressions were identified as having significant correlation with FEV1 changes after LABA treatment: CLN8, PCSK5, and SKP2. For LAMA treatment, four (C1orf115, PRKX, KIAA1618, and RHOQ) were found to be associated with significant changes in FEV1 values. Figure S3A and B shows the microarray results. The linear regression model built for the prediction of FEV1 changes following treatment, using the gene expressions, is shown in . The R2 of the models for LABA and LAMA treatment were 0.158 and 0.404, respectively.
Table 4 Linear regression models for predicting FEV1 changes after LABA and LAMA, using gene expressions
The analysis of damaged-gene scores yielded four and three genes, respectively, for the LABA and LAMA models. EPG5, FNBP4, SCN10A, and SPTBN5 were found to be associated with prediction of LABA response, whereas FBN3, FDFT1, and ZBED6 were found to predict LAMA treatment response (Figure S4A and B). They were used in the prediction models, as shown in (R2=0.318 and 0.199 for the LABA and LAMA treatment, respectively).
Table 5 Linear regression models for predicting FEV1 changes after LABA and LAMA, using damaged-gene scores
Prediction models for long-acting bronchodilator treatment response using gene expressions and damaged-gene scores in addition to clinical traits
The clinical traits, gene expressions, and damaged-gene scores were included together to build the final prediction models for treatment response (). When the clinical traits, gene expressions, and damaged-gene scores were combined together in the prediction model, the predictability was highest (R2=0.512 for LABA and 0.575 for LAMA treatment; both p-values <0.001).
Table 6 Linear regression models for predicting FEV1 changes after LABA and LAMA, using clinical traits, gene expressions, and damaged-gene scores
Discussion
In this multicenter, prospective study, we built prediction models for treatment response to long-acting bronchodilators using the pretreatment traits of the patients, before the treatment was initiated. In total, two clinical traits and seven genes were found to be correlated with the FEV1 changes and were included in the model. The clinical traits alone were not enough to predict the treatment response; however, when gene expressions and damaged-gene scores were added, the predictability increased.
In this study, the efficacy of the treatment, defined as FEV1 changes after bronchodilator treatment, was similar to the results of previous studies.Citation28,Citation29 However, individual treatment response varied. About a quarter of the patients had a response to both bronchodilators, whereas the rest responded only to one or the other, or to none. Heterogeneity in the phenotypes of COPD patients is thought to be one of the reasons for the varied responsiveness to bronchodilator treatment,Citation13 and genetic variations in the β-2 adrenergic receptorsCitation30 and muscarinic receptors may play a role.Citation17 No conclusive measure exists to predict who will respond to one class or the other.
Bronchodilator reversibility was included in both of the final prediction models for the two types of treatment ( and ). It is reasonable to assume that LABA treatment response is partially predicted by BDR, a response to the short-acting β-2 agonist that acts on the same beta receptors to dilate the airways. Although anticholinergics act on different receptors (muscarinic receptors), BDR was also associated with the FEV1 changes in response to the muscarinic antagonist treatment (). Previous reports showed signal interactions between muscarinic and adrenergic receptors, which might explain this.Citation31–Citation33 Despite this theoretical background and the correlation shown in our study, however, bronchodilator reversibility was not enough to predict the treatment responses to long-acting bronchodilators.
Of note, CAT score was statistically significant in predicting the treatment response to a muscarinic antagonist in the multivariate analysis (). The mechanism of the relationship is unknown, because no previous studies have evaluated the relationship between CAT score and the treatment response to anticholinergic agents. One plausible explanation is that one of the items included in the CAT evaluates the amount of phlegm, which is possibly an important indicator of increased cholinergic tone.Citation34 With respect to the symptoms, we further analyzed the relationship between long-acting bronchodilator treatment responses and the changes in the patients’ symptoms, such as cough, dyspnea, sputum, and CAT score. A significant association was found between improvement in FEV1 following the LAMA treatment and symptom improvement (ie, CAT score change); FEV1 change after LABA treatment did not show a significant association with symptom changes (Table S3). This finding hints at an association between CAT score and LAMA treatment response, although the specific mechanisms are yet to be determined.
Our study employed a novel approach by using gene analysis and clinical traits together to build a prediction model for long-acting bronchodilator treatment response. We used peripheral blood to investigate genetic traits. Sampling lung tissue is invasive and peripheral blood has been utilized as a noninvasive alternative.Citation35–Citation38 The rationale for the use of peripheral blood is that COPD is a systemic disease.Citation39 Moreover, some overlapping gene expressions were found between blood and lung tissueCitation35 or alveolar macrophageCitation36 in previous studies. Thus, we hypothesized that gene expressions and changes in the DNA sequencing identified from peripheral blood may reflect those of the lung. The genes found to be highly correlated with FEV1 changes following treatment with bronchodilators in our study were rather unexpected. Although β-2-adrenergic receptor gene (ADRB2), for example, has been identified with genetic variationsCitation18,Citation40,Citation41 and suggested to be one of the markers that partially explain variable responses to a short-acting beta agonist,Citation30 its role as the sole genetic determinant of a response to bronchodilators is not conclusive.Citation42,Citation43 Also, the genetic effects on long-acting bronchodilator treatment responses have not been determined.Citation44 It is difficult to explain the association between the treatment response and the genes found in this study, because the biologic functions of some genes have not been confirmed. In addition, this result might have been derived by chance following the tests in which numerous genes were analyzed at once. Even though the true influence of these genes is unclear, there may be some plausible explanations. For example, SCN10A is known to encode proteins associated with voltage-gated sodium channels that are required in excitable cells including muscles.Citation45 Because bronchial smooth muscles constrict and relax in response to voltage changes, we may assume that a damage in this gene can have a reduced bronchodilator response. RHOQ may have a role in trafficking of CFTR, which is found in epithelial cellsCitation46 and thought to be associated with thickness of mucus and airway clearance. Thus, expression of RHOQ may be related with airway clearance, explaining the association with the treatment response to a LAMA.
Clinical traits such as CAT scores and spirometry results are easy to obtain and are noninvasive. However, clinical traits were not enough to predict the treatment responses to long-acting bronchodilators (R2=0.231 and 0.121, for the LABA and LAMA model, respectively). When only genetic data were used to build the prediction model, the predictability slightly increased. In the final model we built, using the two clinical traits, gene expressions, and damaged-gene scores together, the predictability was highest, at R2=0.512 and 0.575 for the LABA and LAMA model, respectively. Given the recent advances in COPD genetics and the ongoing active research, our method of considering genetic data in addition to clinical traits may be a guide for further studies.
There are some limitations to our study. First, the results might have been influenced by the small number of included patients. However, some variables were demonstrated to be statistically significant, with a reasonable explanation, such as BDR. Second, as mentioned before, it is difficult to explain how the genes found in this study function as predictors of the treatment response. Nevertheless, the genes were found through the use of rigorous methods; professionals performed the RNA microarray and exome sequencing. Also, analytic validation was done to ensure the accuracy of the results. The specific roles of the genes remain a subject for further research. Third, the model did not undergo clinical validation, and further studies with larger numbers of patients are needed.
In spite of the limitations, our study has value in that it is the first study that included the genetic traits and the clinical traits of the patients in building a prediction model for long-acting bronchodilator treatment responses. In addition, the gene analyses were performed by professionals with reliable accuracy, and the data underwent analytic validation. Although how the genes identified in this study are involved in COPD treatment responsiveness remains unknown, the prediction model may provide a guide for future research to incorporate genetic data and clinical traits in predicting the treatment response to long-acting bronchodilators.
Acknowledgments
This work was supported by the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (grant number HI13C1634, HC16C2254). The authors thank Dr Sei Won Lee for critical discussions and support to this work.
Disclosure
The authors report no conflicts of interest in this work.
References
- LozanoRNaghaviMForemanKGlobal and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010Lancet201238098592095212823245604
- Global Initiative for Chronic Obstructive Lung Disease (GOLD)Global Strategy for the Diagnosis, Management and Prevention of COPD2017 Available from: http://goldcopd.org/Accessed October 20, 2017
- TashkinDPCooperCBThe role of long-acting bronchodilators in the management of stable COPDChest2004125124925914718448
- MahlerDADonohueJFBarbeeRAEfficacy of salmeterol xinafoate in the treatment of COPDChest1999115495796510208192
- CasaburiRMahlerDAJonesPWA long-term evaluation of once-daily inhaled tiotropium in chronic obstructive pulmonary diseaseEur Respir J200219221722411866001
- VinckenWvan NoordJAGreefhorstAPImproved health outcomes in patients with COPD during 1 yr’s treatment with tiotropiumEur Respir J200219220921611871363
- SantusPRadovanovicDPaggiaroPWhy use long acting bronchodilators in chronic obstructive lung diseases? An extensive review on formoterol and salmeterolEur J Intern Med201526637938426049917
- OkudanNGokMGokbelHSuerdemMSingle dose of tiotropium improves the 6-minute walk distance in chronic obstructive pulmonary diseaseLung2006184420120417006746
- BrouillardCPepinVMilotJLacasseYMaltaisFEndurance shuttle walking test: responsiveness to salmeterol in COPDEur Respir J200831357958418057052
- BarrRGBourbeauJCamargoCARamFSTiotropium for stable chronic obstructive pulmonary disease: a meta-analysisThorax2006611085486216844726
- JonesPWBarnesNVogelmeierCLawrenceDKramerBEfficacy of indacaterol in the treatment of patients with COPDPrim Care Respir J201120438038821785813
- MiravitllesMSoler-CatalunaJJCalleMSorianoJBTreatment of COPD by clinical phenotypes: putting old evidence into clinical practiceEur Respir J20134161252125623060631
- AndersonDMacneeWTargeted treatment in COPD: a multi-system approach for a multi-system diseaseInt J Chron Obstruct Pulmon Dis2009432133519750192
- AnthonisenNRLindgrenPGTashkinDPKannerREScanlonPDConnettJEBronchodilator response in the lung health study over 11 yearsEur Respir J2005261455115994388
- LehmannSBakkePSEideGEHumerfeltSGulsvikABronchodilator reversibility testing in an adult general population; the importance of smoking and anthropometrical variables on the response to a beta2-agonistPulm Pharmacol Ther200619427228016169762
- SchermerTHeijdraYZadelSFlow and volume responses after routine salbutamol reversibility testing in mild to very severe COPDRespir Med200710161355136217098407
- CherubiniEEspositoMCScozziDGenetic polymorphism of CHRM2 in COPD: clinical significance and therapeutic implicationsJ Cell Physiol201623181745175126633752
- WechslerMELehmanELazarusSCbeta-Adrenergic receptor polymorphisms and response to salmeterolAm J Respir Crit Care Med2006173551952616322642
- BleeckerERMeyersDABaileyWCADRB2 polymorphisms and budesonide/formoterol responses in COPDChest2012142232032822383665
- KimWJOhYMSungJLung function response to 12-week treatment with combined inhalation of long-acting beta2 agonist and glucocorticoid according to ADRB2 polymorphism in patients with chronic obstructive pulmonary diseaseLung2008186638138618758858
- DonohueJFMinimal clinically important differences in COPD lung functionCOPD20052111112417136971
- KumarPHenikoffSNgPCPredicting the effects of coding non-synonymous variants on protein function using the SIFT algorithmNat Protoc2009471073108119561590
- AutonABrooksLDDurbinRMA global reference for human genetic variationNature20155267571687426432245
- NakkenSAlsethIRognesTComputational prediction of the effects of non-synonymous single nucleotide polymorphisms in human DNA repair genesNeuroscience200714541273127917055652
- YuETHadiMZBioinformatic processing to identify single nucleotide polymorphism that potentially affect Ape1 functionMutat Res2011722214014620633698
- AroraSHuwePJSikderRFunctional analysis of rare variants in mismatch repair proteins augments results from computation-based predictive methodsCancer Biol Ther201718751953328494185
- LeeKHBaikSYLeeSYParkCHParkPJKimJHGenome sequence variability predicts drug precautions and withdrawals from the marketPLoS One2016119e016213527690231
- OhnoTWadaSHanadaSSawaguchiHMurakiMTohdaYEfficacy of indacaterol on quality of life and pulmonary function in patients with COPD and inhaler device preferencesInt J Chron Obstruct Pulmon Dis2014910711424489464
- MoitaJBarbaraCCardosoJTiotropium improves FEV1 in patients with COPD irrespective of smoking statusPulm Pharmacol Ther200821114615117693107
- HizawaNMakitaHNasuharaYBeta2-adrenergic receptor genetic polymorphisms and short-term bronchodilator responses in patients with COPDChest200713251485149217890463
- ProskocilBJFryerADBeta2-agonist and anticholinergic drugs in the treatment of lung diseaseProc Am Thorac Soc200524305310 discussion 311–30216267353
- CazzolaMMolimardMThe scientific rationale for combining long-acting beta2-agonists and muscarinic antagonists in COPDPulm Pharmacol Ther201023425726720381630
- KurianNHallCJWilkinsonGFSullivanMTobinABWillarsGBFull and partial agonists of muscarinic M3 receptors reveal single and oscillatory Ca2+ responses by beta 2-adrenoceptorsJ Pharmacol Exp Ther2009330250251219420300
- BelmonteKECholinergic pathways in the lungs and anticholinergic therapy for chronic obstructive pulmonary diseaseProc Am Thorac Soc200524297304 discussion 311–29216267352
- BhattacharyaSTyagiSSrisumaSPeripheral blood gene expression profiles in COPD subjectsJ Clin Bioinform20111112
- PoliskaSCsankyESzantoAChronic obstructive pulmonary disease-specific gene expression signatures of alveolar macrophages as well as peripheral blood monocytes overlap and correlate with lung functionRespiration201181649951021430361
- EdmistonJSArcherKJScianMJGene expression profiling of peripheral blood leukocytes identifies potential novel biomarkers of chronic obstructive pulmonary disease in current and former smokersBiomarkers201015871573020887155
- BahrTMHughesGJArmstrongMPeripheral blood mononuclear cell gene expression in chronic obstructive pulmonary diseaseAm J Respir Cell Mol Biol201349231632323590301
- AgustiASorianoJBCOPD as a systemic diseaseCOPD20085213313818415812
- IsraelEDrazenJMLiggettSBThe effect of polymorphisms of the beta(2)-adrenergic receptor on the response to regular use of albuterol in asthmaAm J Respir Crit Care Med20001621758010903223
- IsraelEChinchilliVMFordJGUse of regularly scheduled albuterol treatment in asthma: genotype-stratified, randomised, placebo-controlled cross-over trialLancet200436494441505151215500895
- HizawaNPharmacogenetics of chronic obstructive pulmonary diseasePharmacogenomics201314101215122523859575
- HershCPPharmacogenetics of chronic obstructive pulmonary disease: challenges and opportunitiesPharmacogenomics201011223724720136362
- CondreayLHuangLHarrisEGenetic effects on treatment response of umeclidinium/vilanterol in chronic obstructive pulmonary diseaseRespir Med201611412312627109822
- GeneCardsSCN10A Gene Available from: http://www.genecards.org/cgi-bin/carddisp.pl?gene=SCN10AAccessed October 20, 2017
- GeneCardsRHOQ Gene Available from: http://www.genecards.org/cgi-bin/carddisp.pl?gene=RHOQ&keywords=rhoqAccessed October 20, 2017