Abstract
Purpose
Chronic obstructive pulmonary disease (COPD) is a worldwide public health challenge due to its high prevalence and related disability and mortality; however, the pathogenesis of COPD remains unclear. In this study, we aimed to identify key proteins involved in the pathogenesis of COPD.
Patients and Methods
We collected lung tissue from three patients with COPD who required thoracic surgery for lung transplantation in the China–Japan Friendship Hospital. Lung tissue from three donors who had no history of lung disease was collected as healthy controls through a whole-body donation program of Peking Union Medical College (China). We conducted a proteomic analysis of the protein expression profiles in the two groups using a combination of high-resolution liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) and quantitative 6-plex tandem mass tag-labeling; these data were validated by Western blot analysis.
Results
A total of 4976 proteins were identified and analyzed, of which 173 were significantly changed (118 downregulated and 55 upregulated). Gene ontology analysis and protein–protein interaction networks demonstrated that the significantly changed proteins, especially downregulated proteins, were involved in platelet and macrophage activation. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD017158.
Conclusion
In our study, GP6, PF4, and THBS1, which are associated with platelet activation and wound healing, were significantly downregulated in COPD patients. These results indicate that patients with COPD are more likely to develop hemostasis disorders, which could impede the repair process of the lung tissues. Moreover, downregulation of CD163, MARCO and VSIG4, which are involved in dysfunction of alveolar macrophages in efferocytosis, may inhibit the resolution of inflammation and contribute to the pathogenesis of COPD.
Introduction
Chronic obstructive pulmonary disease (COPD) is associated with high levels of disability and mortality.Citation1,Citation2 In 2015, the number of individuals with COPD was estimated at 174.5 million worldwide,Citation2 affecting around 99.9 million people (8.6% of the population aged 20 and over) in China.Citation1
The pathogenesis of COPD is complex and heterogeneous, and is not well elucidated.Citation3 Disorders of repair and remodeling of lung tissue damage can contribute to the pathogenesis of COPD.Citation3 Previous studies indicated that macrophages, which are critical for the resolution of inflammation, exhibited dysfunctional responses to infection and had lower phagocytic and bactericidal activity in the lungs of individuals with COPD.Citation4 Some studies have also provided evidence of autoimmunity in patients with COPD. For instance, circulating antibodies against elastin and pulmonary epithelium and endothelium as well as CD4+ T cells which responded to elastin were found in patients with COPD.Citation5 However, the precise pathological mechanisms of COPD remain to be fully clarified.
Proteomics is an important technique that is widely used to identify the key molecules and pathways in a variety of physiological and pathological processes.Citation6 In a recent study focusing on the proteomics of COPD,Citation7 a total of 24 significantly changed proteins were identified in exhaled breath condensate (EBC) samples. Most of these proteins were involved in inflammation. However, in that study, only 257 proteins were identified, and the concentrations of these proteins were low, due to the technological limitations.Citation7 No recent studies have revealed the proteomic profiles of lung tissue of patients with COPD.
In this study, we adopted a tandem mass tag (TMT)-labeled quantitative proteomic approach to the analysis of lung tissues from patients with COPD and healthy donors. Changes in differentially expressed proteins and protein–protein interactions of significantly changed proteins related to COPD were identified using bioinformatics approaches. Our results indicated that hemostasis disorders and failure to resolve inflammation caused by platelet and macrophage dysfunction are critically involved in the pathogenesis of COPD.
Materials and Methods
Reagents
The following reagents and kits were used in this study: urea (GE Healthcare, LC, UK); protease inhibitor cocktail (Roche, BS, CH); Tandem Mass TagTM kits (Thermo Fisher Scientific, NJ, USA); and sequencing grade Trypsin/Lys-C (Promega, WI, USA). The following antibodies were used in this study: anti-VSIG4 (ab56037, Abcam, Cambridge, UK), anti-CD163 (ab182422, Abcam), anti-TIMP3 (ab39184, Abcam), and anti-ACTB (GTX124213, GeneTex, CA, USA).
Sample Collection and Preparation
In this study, we collected lung tissue samples from three patients with COPD who required thoracic surgery for lung transplantation in the China–Japan Friendship Hospital. As healthy controls, lung tissues from three donors who had no history of lung disease were obtained from the Human Brain Bank, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China. All donors provided written informed consent for using the donated body tissue for medical research. The present study was approved by the ethics committee of the Institute of Basic Medical Sciences of the Chinese Academy of Medical Sciences (Approval Number: 009-2014) and the medical ethics committee of China–Japan Friendship Hospital (Approval Number: 2019-106-K74). The lung samples were washed to remove residual blood and cut into small pieces in ice-cold PBS before centrifugation (12,000 rpm, 3 min).
Protein Extraction and TMT-Labeling
Specimens were prepared for TMT-labeling as described previously.Citation8–Citation10 Samples from three patients with COPD were then labeled with TMT-126, TMT-127, and TMT-128, respectively, while the healthy donors were labeled with TMT-129, TMT-130, TMT-131, respectively. The labeled peptides were then mixed for further MS analysis.
HPLC Fractionation
According to a previous report,Citation11 the mixed peptides were analyzed using an UltiMate 3000 UHPLC (Thermo Fisher Scientific, NJ, USA) fitted with an Xbridge BEH300 C18 column (4.6 × 250 mm, 5 µm, 300 Å, Waters). Using a mobile phase A consisting of H2O and a mobile phase B consisting of 98% acetonitrile (both pH 10), the peptides were separated by gradient elution as previously reported.Citation10 A total of 47 fractions were collected at 1.5 min intervals starting at 2 min after the elution was initiated. The fractions were then dried under vacuum and combined into 12 fractions before dissolution in 20 µL 0.1% TFA for LC-MS/MS analysis.
LC-MS/MS Analysis
For LC-MS/MS analysis, peptides were separated by gradient elution using mobile phase A (0.1% formic acid) and mobile phase B (100% acetonitrile and 0.1% formic acid). The elution was conducted over a period of 135 min at a flow rate of 0.3 µL/min. MS data were acquired using an Orbitrap Fusion mass spectrometer in data-dependent acquisition mode. A single full-scan mass spectrum (350–1550 m/z, 120,000 resolution) was obtained followed by 3-s data-dependent MS/MS scans with the ion routing multipole set at 35% and high-normalized collision energy. Data were analyzed using Xcalibur 4.1 software.
Bioinformatics Analysis
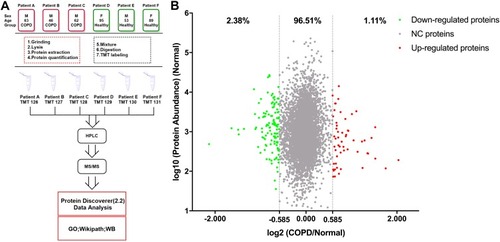
For bioinformatics analysis, Thermo Fisher Scientific Proteome Discover software suite 2.2 was used for LC-MS/MS data analysis with the SEQUEST search engine. The raw MS data files were searched against the UniProt/SwissProt human proteome database (20190604). For each protein, the average abundance of the three patients with COPD (COPD group) and three healthy donors (healthy group) were calculated separately and used for analysis. For significantly changed proteins, the thresholds for downregulation and upregulation were set 0.667 (2−0.584) and 1.50 (20.584), respectively. The PANTHER database (http://www.pantherdb.org/) was used for gene ontology (GO) analysis and protein–protein interaction networks were predicted using the STRING database (http://www.string-db.org) with protein accessions as input and a confidence level threshold of 0.4 (medium confidence). The interaction networks were visualized with Cytoscape 3.6.1 and highly connected clusters were identified using Molecular Complex Detection (MCODE) 1.5.1Citation12 with default parameters (degree cut-off: 2; node score cut-off: 0.2; K-core: 2; max. depth from seed: 100). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repositoryCitation13 with the dataset identifier PXD017158. The workflow of this study is shown in .
Figure 1 The workflow of this study and scatter diagram of identified proteins. (A) Experimental workflow of proteomic profiling of lung tissues. (B) Abundance ratios of proteins are depicted for comparisons of patients with chronic obstructive pulmonary disease (COPD) and healthy donors. Proteins with an abundance ratio <0.667 or >1.50 were defined as significantly changed (SC) proteins. Downregulated proteins are represented as green dots, while upregulated proteins are depicted as red dots. Other proteins are defined as non-significantly changed (NC) proteins and are shown as gray dots.
Western Blot Analysis
Western blot analysis of tissue lysates (10 μg per sample) was performed using standard protocols. Proteins were detected with primary antibodies (1:2000 for TIMP3; 1:1000 for CD163 and 1:200 for VSIG4) and horseradish peroxidase-conjugated secondary antibodies. Protein bands were visualized using ECL reagents and imaged for quantification of band intensity.
Statistical Analysis
Western blot data were analyzed by two-tailed Student’s t-test. P < 0.05 was considered to indicate statistical significance. The statistical figures were drawn with GraphPad Prism 8.0 software (GraphPad, San Diego, CA, USA).
Results
Sample Information
In this study, lung tissue samples were resected from patients with COPD during surgery and from healthy donors through a whole-body donation program. Specimens were divided into two groups: the COPD group (3 males, average age = 57 years; labeled with TMT-126, TMT-127, TMT-128, respectively), and the healthy group (1 male and 2 females, average age = 79 years; labeled with TMT-129, TMT-130, TMT-131, respectively).
Protein Profiles of Lung Tissues from Individuals with COPD and Without COPD Showed Significant Differences in Protein Expression
The LC-MS/MS analysis of TMT-labeled tissue samples from individuals with and without COPD revealed a total of 4976 proteins (unique peptides ≥2, false discovery rate (FDR) ≤0.01, Supplementary Table S1). Using the UniProt mapping tool, 4968 of these proteins were successfully mapped from the UniProt accession profiles following conversion into corresponding Entrez Gene IDs. Comparison of the COPD group with the healthy group revealed 55 (1.11%) upregulated proteins and 118 (2.38%) downregulated proteins (ratio >1.50, or <0.67, ). The significantly changed proteins detected in this study are shown in Supplementary Table S2.
GO and Protein–Protein Interaction Analyses of COPD Suggested Platelet and Macrophage Dysfunction
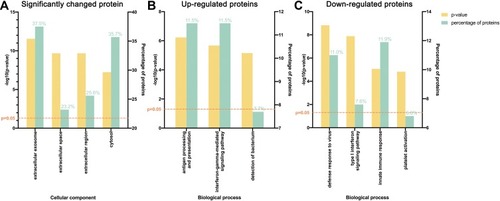
GO analysis was next performed. Significantly changed proteins were classified using PANTHER. Most of these proteins were from extracellular exosomes (37.5%), cytosol (35.7%), and extracellular space (23.2%, ). For the upregulated proteins, the most prominent biological processes included antigen processing and presentation (11.5%) and the interferon-gamma-mediated signaling pathway (11.5%, ), while for downregulated proteins, the most prominent biological processes were innate immune response (11.9%) and defense response to virus (11.0%, ).
Figure 2 Gene ontology (GO) analysis demonstrating protein classification of significantly changed proteins using PANTHER (http://www.pantherdb.org/). (A) All significantly changed proteins are classified according to cellular component. (B) Upregulated and (C) downregulated proteins are classified based on biological process.
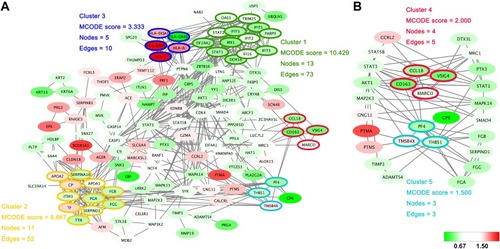
Within the protein–protein interactions predicted among the significantly changed proteins shown in , five highly connected clusters were identified, with MCODE scores ≥1.5 and nodes ≥3. Interestingly, one of these highly connected clusters () was predominantly related to macrophage-mediated clearance of pathogens, containing proteins such as scavenger receptor cysteine-rich type 1 protein M130 (CD163), V-set and immunoglobulin domain-containing protein 4 (VSIG4) and macrophage receptor (MARCO). Another highly connected cluster contained proteins predominantly related to platelet activation, such as platelet factor 4 (PF4), platelet glycoprotein VI (GP6).
Figure 3 Protein–protein interaction networks. (A) The protein–protein interactions of all significantly changed proteins were analyzed with STRING (http://www.string-db.org). The MCODE plugin tool in Cytoscape was used for further analysis of densely connected regions. (B) Two highly connected clusters (Cluster 4 and Cluster 5) and proteins interacting with these clusters are shown. Cluster 4 is mainly related to the function of macrophages, and cluster 5 is closely related to platelet activation. The upregulated proteins are shown with a red background and the downregulated proteins are shown with a green background.
Validation of MS/MS Spectrum Data by Western Blot Analysis
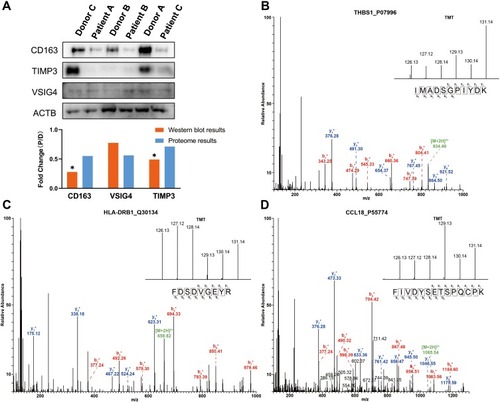
To validate the accuracy of the quantitative proteomic analyses, three proteins (CD163, VSIG4 and TIMP3) involved in platelet and macrophage activation were analyzed by Western blotting. No significant changes in β-actin (ACTB) levels were found between patients and donors, and thus ACTB was chosen as the reference protein. In accordance with the proteomics data, CD163, VSIG4, and metalloproteinase inhibitor 3 (TIMP3) were downregulated in the lung tissues of patients with COPD (). As shown in –, the MS/MS spectra for thrombospondin-1 (THBS1), HLA class II histocompatibility antigen, DRB1 beta chain (HLA-DRB1), and C-C motif chemokine 18 (CCL18) were consistent with the corresponding proteomics data.
Figure 4 Western blot and MS/MS analyses indicating the changes in protein expression. (A) Western blot analyses of CD163, VSIG4 and TIMP3 expression in lung tissues of COPD patients and healthy donors. The results indicate the consistency with the mass spectrometry data. “D” represents healthy donors and “P” represents COPD patients. *P < 0.05 (two-tailed Student’s t-test). (B–D) The representative MS/MS spectrum data of THBS1, HLA-DRB1, and CCL18. The column height of the TMT diagram indicates the relative quantification of the peptide segment in Patients A, B, and C and Donors A, B, and C; 126.13 (TMT-126) for Patient A, 127.12 (TMT-127) for Patient B, 128.14 (TMT-128) for Patient C, 129.13 (TMT-129) for Donor A, 130.14 (TMT-130) for Donor B, and 131.14 (TMT-131) for Donor C.
Discussion
In this proteomics analysis of lung tissues of patients with COPD and healthy donors, we identified 4976 proteins, of which 173 proteins were significantly changed (55 upregulated, 118 downregulated). According to GO classification, most of these significantly changed proteins were from extracellular exosomes and the cytosol. The upregulated proteins were mostly involved in antigen processing and presentation, while the downregulated proteins were mostly involved in innate immune responses and platelet activation. Moreover, the protein–protein interaction analysis indicated that some highly connected clusters were related to pathogen clearance by macrophages and platelet activation.
The pathogenesis of COPD is complex and heterogeneous, and is not well elucidated.Citation3 Disorders of repair and remodeling following lung tissue damage may contribute to the pathogenesis of COPD.Citation3 Normally, the repair process begins with activation of the coagulation system, which initiates the processes required to stop bleeding.Citation14 This is followed by the infiltration of inflammatory immune cells, primarily neutrophils and macrophages, which protect the site of injury from infection and participate in a process that removes dead and damaged tissue.Citation14 Repetitive injury, such as that induced by smoking, elicits a more complex form of tissue repair that combines tissue destruction with scar formation in patients with COPD.Citation15 In patients with COPD, the disorders of repair and remodeling could be a result of dysregulated hemostasis or failure to resolve inflammation.
According to our results, many proteins related to platelet activation, such as GP6, PF4, and THBS1 were downregulated. Furthermore, GO analysis indicated that 6.8% of the downregulated proteins were involved in platelet activation. Platelet GP6 interacts with the exposed collagen, resulting in platelet procoagulant activity and platelet plug formation.Citation16 PF4 is a chemokine that initiates platelet aggregation and also has chemotactic activity for neutrophils and monocytes.Citation17 THBS1, which is involved in cell-to-cell and cell-to-matrix interactions, plays important roles in wound healing, inflammation and angiogenesis.Citation18 GP6, PF4, and THBS1 were all downregulated in patients with COPD, which would cause bleeding disorders, and interrupt repair process.
Once the inflammatory stimulus ceases, resolution of inflammation begins. Macrophages play an important role in elimination of apoptotic neutrophils and epithelial cells (efferocytosis), which prevent secondary necrosis of apoptotic cells with liberation of the proinflammatory cytoplasmic content.Citation3,Citation19 One of the highly connected clusters in the protein–protein interaction networks identified in this study was related to macrophage activation, and proteins in the cluster (eg CD163, MARCO and VSIG4) were mostly downregulated. The GO analysis also indicated that most of the downregulated proteins were involved in innate immune responses.
Alveolar macrophages from COPD patients have been reported to display aberrant efferocytosis. This defect is more obvious in current smokers, possibly due to modification of extracellular macrophage proteins caused by exposure to cigarette smoke;Citation20,Citation21 however, this remains to be confirmed. The resulting failure to remove apoptotic cells in the lungs leads to secondary necrosis and the release of inflammatory mediators that exacerbate lung damage.Citation22
CD163 is involved in the endocytosis of components of damaged cells by macrophages, a process that dampens the inflammatory response.Citation23 Abdullah et al demonstrated that CD163 plays an important role in antioxidant, inflammation-influencing and immunomodulatory control mechanisms in human lung using ex vivo cultured lung tissue.Citation24 Our results indicated that CD163 was significantly downregulated in patients with COPD, which could prevent clearance of apoptotic cells and promote the maintenance of macrophages in a proinflammatory phenotype. It can be speculated that these changes play an important role in the mechanism underlying the chronic inflammatory profile observed in many COPD patients.Citation19
The transmembrane receptor MARCO, which is expressed by alveolar macrophages, is involved in inflammation and pathogen clearance.Citation25 Our results indicated that MARCO was significantly downregulated. Cigarette smoking, which is the leading cause of COPD, has been reported to downregulate MARCO in an effect that might be responsible for impaired control of inflammation.Citation26 Dahl et al also demonstrated that MARCO-knockout mice showed increased pulmonary inflammation and greater lung injury than wild-type mice following exposure to ozone.Citation27
VSIG4, which is expressed by resting macrophages, has been confirmed as a negative regulator of T cell activationCitation28 and plays an inhibitory role in macrophage-mediated inflammation.Citation29 The expression of VSIG4 is restricted to tissue macrophages, including alveolar macrophages.Citation30 Our results indicated that VSIG4 was significantly downregulated. Interestingly, VSIG4 may prevent local inflammation by participating in the essential removal of C3-opsonized apoptotic cells and cell debris.Citation30 In vitro assays have been used to confirm that this biological process is dysfunctional in patients with COPD.Citation31 The significant downregulation of VSIG4 we observed appears to play an important role in the dysfunction of alveolar macrophages in COPD patients.
Conclusion
Our study indicates that the disorders of repairing and remodeling that occur after repetitive injury could contribute greatly to the pathogenesis of COPD, with the hemostasis disorders and failure to resolve inflammation playing the predominant role. Abnormal platelet activation associated with downregulation of GP6, PF4, and THBS1 contributes to the hemostasis disorders. In addition, dysfunction of alveolar macrophages in efferocytosis, related to downregulation of CD163, VSIG4 and MARCO, could inhibit the resolution of inflammation in patients with COPD and contribute to the pathogenesis of COPD.
Disclosure
The authors report no conflicts of interest in this work.
Acknowledgments
The authors thank all donors and their relatives for tissue samples. This study was supported by the National Natural Science Foundation of China (NSFC #81971023), the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (CIFMS, #2018-I2M-1-001) and the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2019-RC-HL-006). Yifan Liu and Haotian Liu are co-first authors for this study.
References
- WangC, XuJ, YangL, et al. Prevalence and risk factors of chronic obstructive pulmonary disease in China (the China Pulmonary Health [CPH] study): a national cross-sectional study. Lancet. 2018;391(10131):1706–1717. doi:10.1016/S0140-6736(18)30841-929650248
- VosT, AllenC, AroraM; Disease GBD, Injury I, Prevalence C. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388(10053):1545–1602. doi:10.1016/S0140-6736(16)31678-627733282
- AgustiA, HoggJC. Update on the pathogenesis of chronic obstructive pulmonary disease. N Engl J Med. 2019;381(13):1248–1256. doi:10.1056/NEJMra190047531553836
- TaylorAE, Finney-HaywardTK, QuintJK, et al. Defective macrophage phagocytosis of bacteria in COPD. Eur Respir J. 2010;35(5):1039–1047. doi:10.1183/09031936.0003670919897561
- CosioMG, SaettaM, AgustiA. Immunologic aspects of chronic obstructive pulmonary disease. N Engl J Med. 2009;360(23):2445–2454. doi:10.1056/NEJMra080475219494220
- ZhuH, SnyderM. ‘Omic’ approaches for unraveling signaling networks. Curr Opin Cell Biol. 2002;14(2):173–179. doi:10.1016/S0955-0674(02)00315-011891116
- SunC, ZhouTY, XieGQ, et al. Proteomics of exhaled breath condensate in stable COPD and non-COPD controls using tandem mass tags (TMTs) quantitative mass spectrometry: a pilot study. J Proteomics. 2019;206:103392.31129269
- XuB, GaoY, ZhanS, et al. Quantitative protein profiling of hippocampus during human aging. Neurobiol Aging. 2016;39:46–56. doi:10.1016/j.neurobiolaging.2015.11.02926923401
- XuB, XiongF, TianR, et al. Temporal lobe in human aging: a quantitative protein profiling study of samples from Chinese Human Brain Bank. Exp Gerontol. 2016;73:31–41. doi:10.1016/j.exger.2015.11.01626631761
- WangX, LiuY, JiaY, et al. Proteome profiling of cerebral vessels in Rhesus Macaques: dysregulation of antioxidant activity and extracellular matrix proteins contributes to cerebrovascular aging in Rhesus Macaques. Front Aging Neurosci. 2019;11:293. doi:10.3389/fnagi.2019.0029331708766
- van UlsenP, KuhnK, PrinzT, et al. Identification of proteins of Neisseria meningitidis induced under iron-limiting conditions using the isobaric tandem mass tag (TMT) labeling approach. Proteomics. 2009;9(7):1771–1781. doi:10.1002/pmic.20080064219288525
- BaderGD, HogueCW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi:10.1186/1471-2105-4-212525261
- MaJ, ChenT, WuS, et al. iProX: an integrated proteome resource. Nucleic Acids Res. 2019;47(D1):D1211–D1217. doi:10.1093/nar/gky86930252093
- KumarV, AbbasAK, FaustoN, RobbinsSL, CotranRS. Robbins and Cotran Pathologic Basis of Disease. 7th ed. Philadelphia: Elsevier Saunders; 2005.
- HoggJC. Pathophysiology of airflow limitation in chronic obstructive pulmonary disease. Lancet. 2004;364(9435):709–721. doi:10.1016/S0140-6736(04)16900-615325838
- Jandrot-PerrusM, BusfieldS, LagrueAH, et al. Cloning, characterization, and functional studies of human and mouse glycoprotein VI: a platelet-specific collagen receptor from the immunoglobulin superfamily. Blood. 2000;96(5):1798–1807. doi:10.1182/blood.V96.5.179810961879
- GuptaSK, HasselT, SinghJP. A potent inhibitor of endothelial-cell proliferation is generated by proteolytic cleavage of the chemokine platelet factor-4. Proc Natl Acad Sci U S A. 1995;92(17):7799–7803. doi:10.1073/pnas.92.17.77997644496
- ChenH, HerndonME, LawlerJ. The cell biology of thrombospondin-1. Matrix Biol. 2000;19(7):597–614. doi:10.1016/S0945-053X(00)00107-411102749
- BelchamberKBR, DonnellyLE. Macrophage dysfunction in respiratory disease. Results Probl Cell Differ. 2017;62:299–313.28455714
- KirkhamPA, SpoonerG, RahmanI, RossiAG. Macrophage phagocytosis of apoptotic neutrophils is compromised by matrix proteins modified by cigarette smoke and lipid peroxidation products. Biochem Biophys Res Commun. 2004;318(1):32–37. doi:10.1016/j.bbrc.2004.04.00315110749
- HodgeS, HodgeG, AhernJ, JersmannH, HolmesM, ReynoldsPN. Smoking alters alveolar macrophage recognition and phagocytic ability: implications in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2007;37(6):748–755. doi:10.1165/rcmb.2007-0025OC17630319
- HodgeS, HodgeG, HolmesM, ReynoldsPN. Increased airway epithelial and T-cell apoptosis in COPD remains despite smoking cessation. Eur Respir J. 2005;25(3):447–454. doi:10.1183/09031936.05.0007760415738287
- KristiansenM, GraversenJH, JacobsenC, et al. Identification of the haemoglobin scavenger receptor. Nature. 2001;409(6817):198–201. doi:10.1038/3505159411196644
- AbdullahM, KahlerD, VockC, et al. Pulmonary haptoglobin and CD163 are functional immunoregulatory elements in the human lung. Respiration. 2012;83(1):61–73. doi:10.1159/00032986821860221
- BinLH, NielsonLD, LiuXQ, MasonRJ, ShuHB. Identification of uteroglobin-related protein 1 and macrophage scavenger receptor with collagenous structure as a lung-specific ligand-receptor pair. J Immunol. 2003;171(2):924–930. doi:10.4049/jimmunol.171.2.92412847263
- BaqirM, ChenCZ, MartinRJ, et al. Cigarette smoke decreases MARCO expression in macrophages: implication in Mycoplasma pneumoniae infection. Respir Med. 2008;102(11):1604–1610. doi:10.1016/j.rmed.2008.05.00218590957
- DahlM, BauerAK, ArredouaniM, et al. Protection against inhaled oxidants through scavenging of oxidized lipids by macrophage receptors MARCO and SR-AI/II. J Clin Invest. 2007;117(3):757–764. doi:10.1172/JCI2996817332894
- VogtL, SchmitzN, KurrerMO, et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J Clin Invest. 2006;116(10):2817–2826. doi:10.1172/JCI2567317016562
- LiJ, DiaoB, GuoS, et al. VSIG4 inhibits proinflammatory macrophage activation by reprogramming mitochondrial pyruvate metabolism. Nat Commun. 2017;8(1):1322. doi:10.1038/s41467-017-01327-429109438
- HelmyKY, KatschkeKJ, GorganiNN, et al. CRIg: a macrophage complement receptor required for phagocytosis of circulating pathogens. Cell. 2006;124(5):915–927. doi:10.1016/j.cell.2005.12.03916530040
- HodgeS, HodgeG, ScicchitanoR, ReynoldsPN, HolmesM. Alveolar macrophages from subjects with chronic obstructive pulmonary disease are deficient in their ability to phagocytose apoptotic airway epithelial cells. Immunol Cell Biol. 2003;81(4):289–296. doi:10.1046/j.1440-1711.2003.t01-1-01170.x12848850
