Abstract
Background and Objectives
Long non-coding RNAs (lncRNAs) play an important role in the pathogenesis of many diseases, including cancer, pulmonary fibrosis and chronic obstructive pulmonary disease (COPD). In this study, we intended to identify the differentially expressed lncRNAs and the role of HOXA cluster antisense RNA 2 (HOXA-AS2) in patients with COPD.
Methods
We analyzed lncRNA profiles of three non-COPD and seven COPD patients’ lungs via microarray and then validated the expression of the top differentially expressed lncRNAs by using real-time polymerase chain reaction (PCR). To identify the mechanism of HOXA-AS2 during COPD pathogenesis and endothelial cell proliferation, we knocked down and overexpressed HOXA-AS2 with siRNA and lentivirus transfection approach in human pulmonary microvascular endothelial cells (HPMECs).
Results
Among 29,150 distinct lncRNA transcripts, 353 lncRNAs were significantly (≥2-fold change and P<0.05) upregulated and 552 were downregulated in COPD patients. The fold change of HOXA-AS2 is 9.32; real-time PCR confirmed that HOXA-AS2 was downregulated in COPD patients. In in vitro experiments, cigarette smoke extract (CSE) treatment reduced the expression of HOXA-AS2 and cell proliferation of HPMECs. Knocking down HOXA-AS2 inhibited HPMECs proliferation and the expression of Notch1 in HPMECs. Overexpressing Notch1 could partly rescue the inhibition of cell viability induced by the silence of HOXA-AS2.
Conclusion
Our results demonstrated that differentially expressed lncRNAs may act as potential molecular biomarkers for the diagnosis of COPD, and HOXA-AS2 was involved in the pathogenesis of COPD by regulating HPMECs proliferation via Notch1, which may provide a new approach for COPD treatment.
Introduction
Chronic obstructive pulmonary disease (COPD) is characterized by progressive and irreversible airflow limitation and associated with an abnormal inflammatory response of the lungs to noxious particles and gases,Citation1 which causes significant morbidity and mortality worldwide and is expected to become the third leading cause of death by 2020.Citation2 It is known that genetic variants and environmental factors have a role in the pathogenesis of this disease. Smoking and/or some other chronic exposure to irritants plays an important role among the environmental factors.Citation3–Citation5 However, only 20% of smokers develop COPD,Citation6 indicating that genetic and epigenetic factors contribute to the susceptibility for this disease.
Protein-coding genes only represent a small part of the human genome (20,687 genes, <2%),Citation7 while a large portion is transcribed into non-coding RNAs (ncRNAs), including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), both of which were originally thought to be non-functional. With the advancement of high-resolution microarray analysis methods, genome-wide sequencing technology or RNA-sequencing, the role of non-coding RNA has generated considerable interest. It was demonstrated that lncRNAs were involved in various cellular functions, including cell proliferation, cell growth, cell differentiation and apoptosis.Citation8–Citation10 The abnormal expression of lncRNAs was revealed in many diseases, including cancer, pulmonary fibrosis, and COPD.Citation11–Citation15 Recently, increasing evidence has shown that HOXA cluster antisense RNA 2 (HOXA-AS2), a lncRNA located between and antisense to the human HOXA3 and HOXA4 genes, is associated with cell survival, proliferation and invasion.Citation16–Citation18 Moreover, it is reported that expression of HOXA-AS2 is related to endothelium inflammation.Citation19 In our previous studies, we found the dysfunction of endothelial cells was involved in the pathogenesis of COPD.Citation20,Citation21 However, whether HOXA-AS2 has a role in the progress of COPD by regulating the function of endothelial cells remains unclear.
In this study, we detected the expression of lncRNAs in the lung tissues of three non-COPD and seven COPD patients by Agilent microarray 3.0 system. We found that HOXA-AS2 was significantly downregulated in COPD patients, and HOXA-AS2 can regulate human pulmonary microvascular endothelial cells (HPMECs) proliferation. Meanwhile, we further investigated the mechanism by which HOXA-AS2 was involved in the pathogenesis of COPD and regulating HPMECs proliferation.
Patients and Methods
Lung Tissue Samples
This study was approved by the Institutional Ethics Committee of the Second Xiangya Hospital of Central South University (2,016,032) and was performed in accordance with the ethical standards established in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. All the patients provided informed consent. COPD patients (n=7) and non-COPD patients (n=3) were diagnosed as spontaneous pneumothorax and required pulmonary segmentectomy or lobectomy. The diagnosis of COPD was based on the Global Initiative for Chronic Obstructive Lung Disease (GOLD) 2013.Citation22 All the patients were in a clinically stable state without pulmonary infection in the last 4 weeks. Patients with other chronic pulmonary diseases, such as obstructive atelectasis and asthma, lung cancer, and other systemic diseases, including diabetes, cardiovascular diseases, renal diseases, hepatic diseases or mental diseases, were excluded. Lung tissue samples were immediately frozen in liquid nitrogen for further experiments, or fixed in 4% formaldehyde and cut into 3.5-mm-thick sections to stain with hematoxylin and eosin (HE).
Arraystar lncRNA Array
Lung tissue RNA was isolated using Trizol reagent (Life Technologies Corporation, CA, USA), and RNA quality was determined. RNA labeling reaction was performed using a Quick Amp Labeling Kit, One-Color (p/n 5190–0442, Agilent Technologies, CA, USA) and purified by RNeasy Mini Kit (p/n 74,104, Qiagen, Hilden, Germany). Then, labeled/amplified cRNA was hybridized using Agilent Gene Expression Hybridization Kit (p/n 5188–5242, Agilent), and then the microarray was washed. We used Agilent Microarray Scanner (p/n G2565BA, Agilent) and Agilent Feature Extraction to scan and extract data. (Aksomics Inc., Shanghai, China)
Cell Culture
HPMECs were purchased from ScienCells Research Laboratory (CA, USA) and were cultured in endothelial cell medium (ECM, ScienCells Research Laboratory, CA, USA) supplemented with 5% fetal bovine serum (FBS), 1% endothelial cell growth factor and penicillin (100 IU/mL)/streptomycin (50 μg/mL) at 37°C in a humidified incubator, containing 5% CO2. Only cells at passages 3 to 4 were used in experiments.
CSE Preparation
CSE was prepared as described previously.Citation20 Briefly, one commercial cigarette (Furong, Changde Cigarette Company, Hunan, China) without filter was burned, and the smoke passed through 25 mL of basic ECM without FBS and endothelial cell growth factor by a vacuum pump. Then, the smoke extract was filtered through a 0.22-μm pore-size filter. The PH of the solution was adjusted to 7.2–7.4. The product was regarded as 100% CSE solution and subsequently diluted with complete ECM to obtain final concentrations of 0.5%, 1%, 2.5%, and 5% in in vitro study.
Cell Transfection
Using Lipofectamine 2000 (Invitrogen, CA, USA) according to the manufacturer’s instructions, HOXA-AS2 siRNA and respective negative control (NC) siRNA (RiboBio Corporation, Guangzhou, China), Notch1 overexpression plasmid vectors and empty vectors (GeneChem, Shanghai, China) were transfected into endothelial cells. For HOXA-AS2 overexpression lentivectors (GeneChem, Shanghai, China) infection, firstly we co-cultured the lentivectors with different cell density in different medium with or without Polybrene or ENi.S.(Enhanced Infection Solution) to find the proper multiplicity of infection (MOI) for lentivectors. Then, the selected condition, including cell seeding density and the most effective conditioned medium, was used in the subsequent infection experiments. The representative images of cell transduction efficiency by lentivectors were shown in Supplementary Material: Figure S1.
Cell Proliferation Assay
The cells were seeded into 96 wells, and treated with the HOXA-AS2 siRNA, or CSE at different concentrations of 0, 0.5%, 1%, 2.5%, and 5%, or 10 mM of Notch1 inhibitor DAPT (GSI-IX, gamma-Secretase Inhibitor IX, ApexBio Technology, TX, USA) for 24h. Then, cell proliferation was carried out by Cell Counting Kit-8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s protocol. In brief, after treatment, the culture medium was aspirated and changed into fresh medium. Then, 10 μL of CCK-8 reagent was added into each well and incubated at 37°C for 4h. The absorbance at 450nm was measured by a microplate reader, which determined the survival rate of endothelial cells.
Real-Time Polymerase Chain Reaction (Real-Time PCR)
Total RNA was extracted from lung tissues, and cells were cultured using Trizol reagent (Life Technologies Corporation, CA, USA). RNA was reverse transcribed using RevertAid™ First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA). Then, real-time PCR was performed with UltraSYBR Mixture (ComWin Biotech Corporation, Beijing, China) following the manufacturer’s protocol in PIKO REAL 96 PCR System (Thermo Fisher Scientific, USA). The sequence of primers is shown in the Supplementary Material: Table S1. We used GAPDH as the internal loading control and calculated the relative expression of lncRNA or mRNA by 2−∆∆CT method. Each PCR analysis was done in triplicate.
Western Blot
Total protein was extracted from cells by RIPA lysis (Beyotime Institute of Biotechnology, Jiangsu, China) containing 1% protease/phosphatase inhibitor cocktail (ComWin Biotech Corporation, Beijing, China) and then measured by Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, USA) to determine the protein concentration. Protein lysates were loaded and separated in 4–20% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel (Sigma, USA) and transferred to polyvinylidene difluoride (PVDF) membranes after electrophoresis (EMD Millipore, Billerica, MA, USA). After blocking with 5% nonfat dry milk for 1 h at room temperature, the membranes were washed and incubated with Notch1 and glyceraldehyde 3-phosphate dehydrogenase (GADPH) primary antibody (20,687-1-AP and 10,494-1-AP, Proteintech Group Incorporation, IL, USA) in dilutions of 1:1000 and 1:3000 at 4°C overnight. The next day, the membranes were washed and incubated with horseradish peroxidase-labeled secondary antibody (SA00001-2, Proteintech Group Incorporation, IL, USA) at a 1:5000 dilution. Then, the antibody-labeled bands were detected by an enhanced chemiluminescence detection system (ECL, BestBio Pharmacia Biotech). The results were quantified with Quantity One Analysis Software (Bio-Rad Laboratories, Hercules, CA, 210 USA).
Statistical Analysis
Statistical analysis of data was conducted by SPSS 22.0 (SPSS Incorporation, IL, USA) and GraphPad Prism 6.0 (GraphPad Software, CA, USA). One-way analysis of variance (ANOVA) was used to analyze the difference between two groups and the least significant difference (LSD) post hoc test was performed to compare the differences between multiple groups. There was statistical significance when p-values were less than 0.05.
Results
Overview of the lncRNA and mRNA Profiles in Lung Tissues of COPD
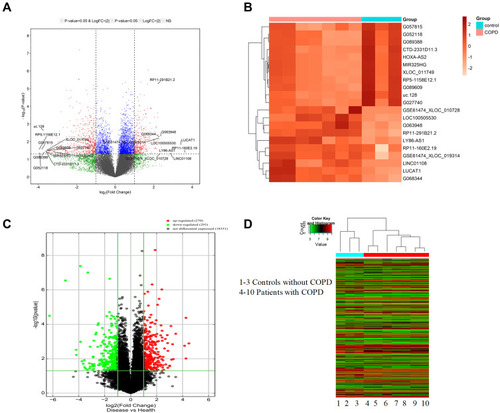
Three non-COPD and seven COPD patients were selected for this study. The basic characteristics are shown in . The hematoxylin-eosin (HE) staining of COPD and non-COPD lung tissues were performed (Supplementary Material: Figure S2). LncRNA array was used to measure the differentially expressed lncRNAs in COPD and non-COPD lung tissues. Twenty-five thousand six hundred and six distinct lncRNAs transcripts were detected in the lung tissues of all subjects. In COPD patients, 353 lncRNAs were significantly (≥2-fold change and P<0.05) up-regulated, and 552 were downregulated when compared to patients without COPD (). RP11-160E2.19, LUCAT1, LINC01108, LY86-AS1, and GO63948 were the top five upregulated lncRNAs with the fold change ranging from 4.67 to 12.23. RP5-1158E12.1, G052118, G057815, Uc.128-, and G089388 were the top five downregulated lncRNAs with the fold change ranging from 12.92 to 15.35 (). Eighteen thousand eight hundred and ninety-four distinct mRNA transcripts were detected in the lung tissues of all subjects. In COPD patients, 270 mRNAs were significantly (≥2-fold change and P<0.05) upregulated and 293 were downregulated when compared to patients without COPD ().
Table 1 Clinical Characteristics of the Subjects Recruited for the Microarray
Figure 1 The differential expression of lncRNAs and mRNA in COPD and non-COPD lung tissues. (A) The volcano plot of differential expressed lncRNAs in COPD compared to non-COPD lung tissues. The expressions of lncRNAs were presented in red (≥2-fold change and P<0.05), green (≥2-fold change but P≥0.05), blue (≤2-fold change and P<0.05), and gray (NS). (B) Hierarchical clustering showed the top differential expression of lncRNAs in COPD (red bar) and non-COPD (blue bar) lung tissues. The expression was displayed on a scale from light (low) to deep (high). (C) The volcano plot of differentially expressed mRNAs in COPD compared to non-COPD lung tissues. The expression of upregulated mRNAs was presented in red (≥2-fold change and P<0.05), downregulated in green (≥2-fold change and P<0.05) and gray (NS). (D) The hierarchical clustering of all differential expressed mRNAs in COPD (red bar) compared to non-COPD (blue bar) lung tissues. The expression was displayed on a scale from green (low) to red (high).
Real-Time PCR Validation
We selected the top five upregulated lncRNAs and six downregulated lncRNAs based on the fold change for confirmation of expression in another two sets of lung tissue samples using real-time PCR, which includes RP11-160E2.19, LUCAT1, LINC01108, LY86-AS1, GO63948, RP5-1158E12.1, G052118, G057815, Uc.128-, G089388, and HOXA-AS2. The basic characteristics of the patients are seen in Supplementary Material: Table S2. The results indicated 63.64% consistency between the microarray data and real-time PCR results. RP11-160E2.19, LUCAT1, GO63948, RP5-1158E12.1, G052118, uc.128-, and HOXA-AS2 were confirmed to change significantly between COPD patients and non-COPD patients ().
Table 2 The Verification of Microarray Analysis Selected from Different Expressed Long Non-Coding RNAs in the List
Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis and Gene Ontology Enrichment Analysis
Pathway analysis demonstrated that there were several enrichment-related pathways. It included 34 upregulated pathways (the top ten up-regulated pathways are shown in ), such as the cytokine-cytokine receptor interaction, chemokine signaling pathway, Toll-like receptor signaling pathway, and NF-kappa B signaling pathway. Ten downregulated pathways () included protein digestion and absorption, steroid hormone biosynthesis, retinol metabolism, and drug metabolism-cytochrome P450.
Figure 2 KEGG pathway analysis and GO enrichment analysis of differentially expressed mRNAs. (A and B) The significantly upregulated (A) and downregulated (B) pathways in COPD vs non-COPD patients. The P-value (EASE-score, Fisher’s P-value, or hypergeometric P-value) denotes the significance of the pathway correlated to the conditions. The lower the P-value, more significant is the pathway (the recommended P-value cutoff is 0.05). (C and D) GO analysis about the top ten counts of enrichment score in the biological process. Up-regulated (C) and down-regulated (D) in the biological process. (E and F) GO analysis of the top ten counts of enrichment score in the biological process. Up-regulated (E) and down-regulated (F) gene molecular function. (G and H) GO analysis of the top ten counts of enrichment score of up-regulated (G) and down-regulated (H) cellular component.
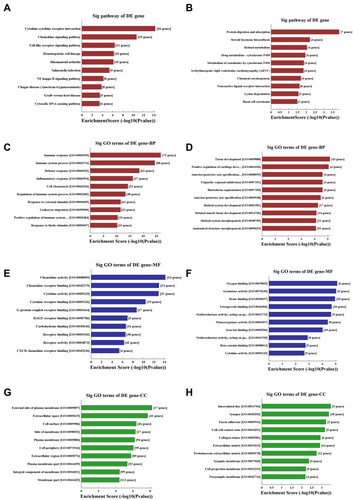
Gene ontology (GO) analysis was performed to determine the gene product enrichment (http://www.geneontology.org). Three domains in ontology were analyzed: biological process (), cellular component (), and molecular function (). For the biological process, there were 526 upregulated and 401 downregulated GO items between the COPD and non-COPD groups. Immune response and tissue development had the highest enrichment score value of the significant enrichment terms.
CSE Treatment Inhibited HPMECs Proliferation and Reduced the Expression of HOXA-AS2
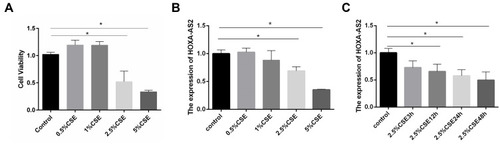
HPMECs were exposed to CSE at different concentrations of 0.0%, 0.5%, 1.0%, 2.5%, and 5.0% for 24 h. CCK-8 assay was then performed to examine cell viability. The results demonstrated that at the concentration of 2.5% CSE, both the cell viability and the expression level of HOXA-AS2 were decreased significantly (). Then, we explored the optimum timepoint for the experiment. The expression of HOXA-AS2 decreased significantly at 12 h, 24 h, and 48 h (). CSE at a concentration of 2.5% was chosen for the following experiment.
Figure 3 CSE treatment inhibited the proliferation of HPMECs and reduced HOXA-AS2 expression. (A) HPMECs were treated with different concentrations of CSE, including 0, 0.5%, 1.0%, 2.5%, and 5.0% for 24 h, and the proliferation of HPMECs was measured by CCK-8 assay. (B) HPMECs was exposed to CSE of different concentrations (0, 0.5%, 1.0%, 2.5%, and 5.0%) for 24 h, and the relative expression levels of HOXA-AS2 was measured by real-time-PCR. At the concentrations of 2.5% and 5% CSE, the cell viability decreased significantly. (C) HPMECs were treated with 2.5% CSE for different amounts of time, and the relative expression levels of HOXA-AS2 were measured by real-time PCR. The expression of HOXA-AS2 decreased significantly as the treatment time extended. Data are presented as mean ± SEM. *P < 0.05. We used GAPDH as the internal loading control and calculated the relative expression of lncRNA by 2−∆∆CT method. All experiments were performed independently at least 3 times.
Effects of HOXA-AS2 on the HPMECs Proliferation
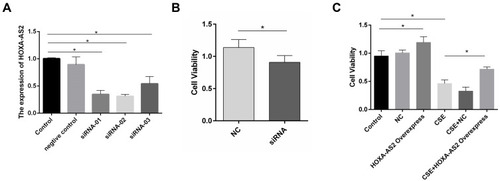
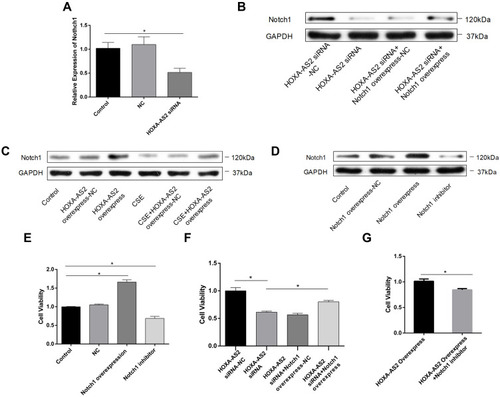
To examine the effect of HOXA-AS2 on cell proliferation, silencing and overexpression approaches were used. We knocked down HOXA-AS2 in HPMECs by transfecting with three kinds of siRNA and found that all the siRNAs caused a significant decrease in HOXA-AS2 expression (). Because siRNA2 transfection had more profound effects on suppressing HOXA-AS2 expression than the other two siRNAs, we chose siRNA2 for the following experiment. The cell viability was significantly inhibited after being transfected with siRNA for 24 h (). To further clarify the role of HOXA-AS2 in regulating cell viability, we overexpressed HOXA-AS2 in HPMECs via transfecting with lentivirus and found that overexpression of HOXA-AS2 could alleviate cell viability inhibition induced by CSE treatment ().
Figure 4 HOXA-AS2 promoted the proliferation of HPMECs. (A) Three kinds of siRNAs were transfected into HPMECs, and the relative expression levels of HOXA-AS2 were measured by real-time PCR. All the siRNAs could cause significant decrease in HOXA-AS2 expression. (B) HPMECs were transfected with negative control (NC) and HOXA-AS2 siRNA for 24 h, and cell viability was measured by CCK-8 assay. This showed that the silence of HOXA-AS2 decreased the proliferation of HPMECs. (C) After transfection with HOXA-AS2 overexpression lentiviral vectors and empty lentiviral vectors, HPMECs were incubated with 2.5% CSE for 24 h. Then, the cell viability was measured by CCK-8 assay. The results suggested that HOXA-AS2 promoted the viability of HPMECs and partly rescued the damage of cell proliferation induced by CSE. All data are presented as mean ± SEM. *P < 0.05. We used GAPDH as the internal loading control and calculated the relative expression of lncRNA by 2−∆∆CT method. All experiments were performed independently at least 3 times.
HOXA-AS2 Regulated HPMECs Proliferation via Notch1
To investigate the mechanisms by which HOXA-AS2 regulates cell proliferation, we used real-time PCR to detect the expression of some important genes related to endothelial proliferation, including Notch1, Fli1, Wnt3a, and vascular endothelial growth factor (VEGF). We found that only Notch 1 was significantly decreased after knocking down HOXA-AS2 (). The other three genes did not present significant differences between the control and HOXA-AS2 siRNA group (Supplementary Material: Figure S3). Western blot also confirmed that knocking down HOXA-AS2 downregulated the expression of Notch1 following the same trend (). In addition, we found that CSE treatment could decrease the expression of Notch1, and it could be partly reversed by overexpressing HOXA-AS2 (). We further confirmed the function of Notch1 in regulating cell proliferation. Notch1 inhibitor and Notch1 overexpression plasmid were used to investigate the functional role of Notch1 in HPMECs. Our results demonstrated that Notch1 inhibitor decreased the expression of Notch1, while Notch1 overexpression could increase the expression of Notch1 (). Importantly, the inhibition of Notch1 decreased cell viability significantly, whereas overexpression of Notch1 could promote the proliferation of HPMECs, suggesting that Notch1 was involved in cell cycle regulation (). To further address the role of Notch1 in HOXA-AS2 regulating cell proliferation, we overexpressed Notch1 in HPMECs after transfecting with HOXA-AS2 siRNA for 24 h. This showed that overexpression of Notch1 could partly rescue the cell viability inhibition induced by the silence of HOXA-AS2 (). Notch1 inhibitor could reduce the cell viability promoted by overexpressing HOXA-AS2 ().
Figure 5 HOXA-AS2 promote HPMECs proliferation through regulating Notch1. (A) HOXA-AS2 siRNAs were transfected into HPMECs for 24 h, and the relative expression levels of Notch1 were measured by real-time PCR. Notch1 mRNA decreased in HOXA-AS2 knocking down HPMECs. (B–D) Western blot was used to analyze the expression of Notch1 protein after transfection with HOXA-AS2 siRNA, overexpressing HOXA-AS2 lentivirus, Notch1 overexpression plasmid or Notch1 inhibitor gamma-Secretase Inhibitor IX (GSI-IX, DAPT 10 mM for 24 h) in HPMECs. (E–G) Cell Counting Kit-8 (CCK-8) assay was used to measure HPMECs viability after treatment. All data are presented as mean ± SEM. *P < 0.05. All experiments were performed independently at least 3 times.
Discussion
The present study showed that lncRNA HOXA-AS2 was significantly downregulated in COPD lung tissues and CSE-treated HPMECs. The expression of HOXA-AS2 showed a dose- and time-dependent decrease in HPMECs when exposed to CSE. We also found that HOXA-AS2 promoted proliferation of HPMECs. Moreover, Notch1 was found to be a downstream molecule of HOXA-AS2, and overexpressing Notch1 protected HPMECs against the damage of cell survival induced by CSE and HOXA-AS2 silencing. Taken together, our study demonstrated that HOXA-AS2 was involved in preservation of endothelial cell viability during the pathogenesis of COPD through regulating Notch1. Our findings strongly suggest the importance of lncRNAs in COPD pathogenesis and likely provide a new target in COPD treatment.
Over the past decades, lncRNAs have been proven to be essential regulators of transcription and translation by their ability to interact with DNAs, RNAs, and proteins.Citation23–Citation25 Moreover, the dysregulation of lncRNAs is associated with pathological processes of different diseases, such as cardiovascular diseases, metabolic diseases, and cancer.Citation26–Citation28 However, the role that lncRNAs plays in COPD pathogenesis remains to be determined. To address this problem, we used microarray analysis to study the differential expression of lncRNAs in COPD and non-COPD lung tissues. Three hundred fifty-three lncRNAs were upregulated, and 552 lncRNAs were downregulated in COPD lung tissues compared to non-COPD tissues. These results suggested that there were significant changes in lncRNA profile in COPD pathological processes. Also, we found that hundreds of mRNAs were significantly differentially expressed in COPD lungs. The GO and pathway analyses showed that lncRNAs was involved in various signaling pathways and metabolic process, such as the NF-kappa B signaling pathway, Wnt/β-catenin signaling pathway, inflammatory response, and immune system. Early studies have shown that NF-kappa B plays an important role in COPD pathogenesis through regulating several inflammatory mediators that are essential to the development of disease.Citation29 The Wnt/β-catenin signaling pathway has also been widely accepted to be related to the destruction of parenchymal tissue and emphysema repair.Citation30 In addition, recent studies have uncovered that immune response has a potential role in COPD pathogenesis.Citation31 Therefore, our study supported that lncRNAs could be involved in COPD progression through various signaling pathways.
Furthermore, the real-time PCR validation showed that lncRNA HOXA-AS2 was significantly downregulated in COPD lung tissue. Recent studies have demonstrated that HOXA-AS2 promotes cell proliferation, migration, and invasion in various human cancers, such as breast cancer, bladder cancer, and osteosarcoma.Citation16,Citation17,Citation32 Zhu et alCitation19 reported that inhibiting HOXA-AS2 could activate the nuclear factor (NF)-κB signaling pathway and induce subsequent inflammatory response in endothelial cells. Endothelium dysfunction is one of the most essential factors contributing to COPD pathogenesis, which has been confirmed by our previous studies.Citation20,Citation21,Citation33 Given the importance of endothelial cells in COPD, we detected the role of HOXA-AS2 in pulmonary endothelial cells. The results showed that the expression of HOXA-AS2 was decreased in HPMECs when exposed to CSE, in dose- and time-dependent reduction. These results revealed that HOXA-AS2 expression within HPMEC level is regulated by CSE stimuli. Mechanistically, HOXA-AS2 silencing suppressed endothelial cell survival, while overexpressing HOXA-AS2 attenuates the viability damage of HPMECs induced by CSE. These results all suggested that HOXA-AS2 could promote the viability of HPMECs, but the mechanisms which were involved in the process needed to be further investigated.
To explore the mechanism of HOXA-AS2 on regulating the proliferation of HPMECs, we searched the online database AnnoLnc (http://annolnc.cbi.pku.edu.cn/) to determine the HOXA-AS2 target protein. Unfortunately, there was no predictional co-expressed protein that was matched in differentially expressed mRNA in microarray analyses. Then, we tried to identify some other important genes related to endothelial proliferation, such as Notch1, Fli1, Wnt3a, and VEGF. The results showed that only Notch 1 was significantly decreased after knocking down HOXA-AS2 in HPMECs. Notch1 has been widely accepted to have significant effects on cell fate determination, including cell proliferation, differentiation, and cell apoptosis.Citation34 When Notch1 is activated by ligands located on the cell surface, Notch1 undergoes intra-membrane proteolysis, then it releases an intracellular region which induces the downstream signaling cascade.Citation35 Our previous study reported that Notch1 was downregulated in HPMECs when exposed to CSE both in vivo and in vitro.Citation20 Concordantly with these early studies, we found that Notch1 overexpression promoted endothelial cell proliferation, whereas NOTCH1 inhibitor decreased cell viability. Interestingly, the expression of Notch1 decreased after knocking down HOXA-AS2 and increased in endothelial cells with HOXA-AS2 overexpression. These data suggested that Notch1 mediates HOXA-AS2-dependent proliferation of HPMECs.
Our study has potential limitations. First, in this study, we verified the top five upregulated lncRNAs, five downregulated lncRNAs, and HOXA-AS2. However, there were hundreds of lncRNAs differentially expressed in COPD lung tissues, and these lncRNAs may also have significant effects in COPD pathogenesis. Second, the present study demonstrated that HOXA-AS2 attenuated the damage of cell viability induced by CSE and promoted proliferation of HPMECs via Notch1. But the mechanism by how HOXA-AS2 regulates Notch1 remains unknown. Third, there was no evidence about the effects of HOXA-AS2 on endothelial cells in an animal model in our study, but the analysis of human lung tissues and thorough study of HOXA-AS2 in endothelial cells can provide strong background information on animal study.
Conclusions
In summary, our study revealed the differential expression level of lncRNAs between COPD and non-COPD lung tissues through microarray analysis. GO and pathway analyses could help us explore and predict the function of certain lncRNAs. The results also suggested that HOXA-AS2 promotes cell proliferation and attenuates the damage of cell viability induced by CSE exposure through upregulation of Notch1. Our findings not only highlight an important role of HOXA-AS2 in the proliferation of HPMECs, but also provide novel insights into searching effective therapeutic targets.
Abbreviations
COPD, chronic obstructive pulmonary disease; CSE, cigarette smoke extract; ECM, endothelial cell media; FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; GOLD, Global Initiative for Chronic Obstructive Lung Disease; HOXA-AS2, HOXA cluster antisense RNA 2; HPMEC, human pulmonary microvascular endothelial cell; LncRNA, long non-coding RNA; PCR, polymerase chain reaction.
Data Sharing Statement
The authors confirm that the data supporting the findings of this study are available within the article. The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request. The authors will not share any individual de-identified participant data or other relevant study documents.
Ethics Approval and Consent to Participate
This research was approved by the Institutional Ethics Committee of the Second Xiangya Hospital of Central South University (2,016,032), and all subjects provided written informed consent to participate in the study.
Author Contributions
PC, AZ, and YYZ contributed to the study conceptualization; ZZ, YYZ, and AZ provided patient lung tissues and clinical information. AZ, ZY, and YZZ contributed to data analysis. AY and YZ performed PCR validation. YYZ and YZ performed the cell experiments. JD and ZZ analyzed the CCK-8 and Western blot data. PC, AZ, and YYZ contributed to funding acquisition. AZ and YYZ contributed to the drafting of the manuscript. PC and SC contributed to critically revising the manuscript for important intellectual content. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
All authors declare no conflicts of interest for this work.
Acknowledgments
The authors thank all the patients who participated in the study. Thank Dr. Zong Dandan (Department of Respiratory and Critical Care Medicine, the Second Xiangya Hospital, Central South University, Changsha, Hunan 410011, China) for her help in providing NOTCH1 overexpression plasmid vectors and thanks Dr. Guo Peipei (Pulmonary, Critical Care and Sleep Medicine, Icahn School of Medicine at Mount Sinai, New York 11766, USA) for helping us check the revised manuscript carefully to make sure the language was accurate and clear enough.
References
- PauwelsRA, BuistAS, CalverleyPM, JenkinsCR, HurdSS, CommitteeGS. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) workshop summary. Am J Respir Crit Care Med. 2001;163(5):1256–1276. doi:10.1164/ajrccm.163.5.210103911316667
- World Health Organization. World health statistics 2008. Available from: http://www.who.int/whosis/whostat/EN_WHS08_Full.pdf. Accessed 520, 2008.
- EzzatiM. Indoor air pollution and health in developing countries. Lancet. 2005;366(9480):104–106. doi:10.1016/S0140-6736(05)66845-616005317
- LiLJ, GaoLB, LvML, et al. Association between SNPs in pre-miRNA and risk of chronic obstructive pulmonary disease. Clin Biochem. 2011;44(10–11):813–816. doi:10.1016/j.clinbiochem.2011.04.02121565178
- TrupinL, EarnestG, San PedroM, et al. The occupational burden of chronic obstructive pulmonary disease. Eur Respir J. 2003;22(3):462–469. doi:10.1183/09031936.03.0009420314516136
- PauwelsRA, RabeKF. Burden and clinical features of chronic obstructive pulmonary disease (COPD). Lancet. 2004;364(9434):613–620. doi:10.1016/S0140-6736(04)16855-415313363
- ConsortiumEP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74.22955616
- ZhangL, ZhouC, QinQ, LiuZ, LiP. LncRNA LEF1-AS1 regulates the migration and proliferation of vascular smooth muscle cells by targeting miR-544a/PTEN axis. J Cell Biochem. 2019;120(9):14670–14678.31016789
- HuangT, WangJ, ZhouY, ZhaoY, HangD, CaoY. LncRNA CASC2 is upregulated in osteoarthritis and participates in the regulation of IL-17 expression and chondrocyte proliferation and apoptosis. Biosci Rep. 2019;39(5). doi:10.1042/BSR20182454
- DelasMJ, JacksonBT, KovacevicT, et al. lncRNA Spehd regulates hematopoietic stem and progenitor cells and is required for multilineage differentiation. Cell Rep. 2019;27(3):719–729 e716. doi:10.1016/j.celrep.2019.03.08030995471
- ZhaoQ, ZhaoS, LiJ, et al. TCF7L2 activated HOXA-AS2 decreased the glucocorticoid sensitivity in acute lymphoblastic leukemia through regulating HOXA3/EGFR/Ras/Raf/MEK/ERK pathway. Biomed Pharmacother. 2019;109:1640–1649. doi:10.1016/j.biopha.2018.10.04630551418
- WangX, ChengZ, DaiL, et al. Knockdown of lncRNA H19 represses the progress of pulmonary fibrosis through the TGF-beta/Smad3 pathway by regulating miR-140. Mol Cell Biol. 2019;39(12). doi:10.1128/MCB.00143-19.
- SchattinA, ArnerR, GennaroF, de BruinED. Adaptations of prefrontal brain activity, executive functions, and gait in healthy elderly following exergame and balance training: a randomized-controlled study. Front Aging Neurosci. 2016;8:278. doi:10.3389/fnagi.2016.0027827932975
- GuC, LiY, LiuJ, et al. LncRNAmediated SIRT1/FoxO3a and SIRT1/p53 signaling pathways regulate type II alveolar epithelial cell senescence in patients with chronic obstructive pulmonary disease. Mol Med Rep. 2017;15(5):3129–3134. doi:10.3892/mmr.2017.636728339038
- ThaiP, StattS, ChenCH, LiangE, CampbellC, WuR. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol. 2013;49(2):204–211. doi:10.1165/rcmb.2013-0159RC23672216
- WangF, WuD, ChenJ, et al. Long non-coding RNA HOXA-AS2 promotes the migration, invasion and stemness of bladder cancer via regulating miR-125b/Smad2 axis. Exp Cell Res. 2019;375(1):1–10. doi:10.1016/j.yexcr.2018.11.005
- WangY, ZhangR, ChengG, XuR, HanX. Long non-coding RNA HOXA-AS2 promotes migration and invasion by acting as a ceRNA of miR-520c-3p in osteosarcoma cells. Cell Cycle. 2018;17(13):1637–1648. doi:10.1080/15384101.2018.148917430081707
- LianY, LiZ, FanY, et al. The lncRNA-HOXA-AS2/EZH2/LSD1 oncogene complex promotes cell proliferation in pancreatic cancer. Am J Transl Res. 2017;9(12):5496–5506.29312501
- ZhuX, LiuY, YuJ, et al. LncRNA HOXA-AS2 represses endothelium inflammation by regulating the activity of NF-kappaB signaling. Atherosclerosis. 2019;281:38–46. doi:10.1016/j.atherosclerosis.2018.12.01230658190
- ZongD, LiJ, CaiS, et al. Notch1 regulates endothelial apoptosis via the ERK pathway in chronic obstructive pulmonary disease. Am J Physiol Cell Physiol. 2018;315(3):C330–C340. doi:10.1152/ajpcell.00182.201729874112
- LongYJ, LiuXP, ChenSS, ZongDD, ChenY, ChenP. miR-34a is involved in CSE-induced apoptosis of human pulmonary microvascular endothelial cells by targeting Notch-1 receptor protein. Respir Res. 2018;19(1):21. doi:10.1186/s12931-018-0722-229373969
- Global Initiative for Chronic Obstructive Lung Disease. Global strategy for diagnosis, management and prevention of chronic obstructive lung disease (updated 2013). Available from: http://goldcopd.org/. Accessed 215, 2013.
- De SmetEG, MestdaghP, VandesompeleJ, BrusselleGG, BrackeKR. Non-coding RNAs in the pathogenesis of COPD. Thorax. 2015;70(8):782–791. doi:10.1136/thoraxjnl-2014-20656025995155
- Soares Do AmaralN, CruzEMN, de Melo MaiaB, Malagoli RochaR. Noncoding RNA profiles in tobacco- and alcohol-associated diseases. Genes (Basel). 2016;8(1):6. doi:10.3390/genes8010006
- GeislerS, CollerJ. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. doi:10.1038/nrm367924105322
- UchidaS, DimmelerS. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015;116(4):737–750. doi:10.1161/CIRCRESAHA.116.30252125677520
- FengSD, YangJH, YaoCH, et al. Potential regulatory mechanisms of lncRNA in diabetes and its complications. Biochem Cell Biol. 2017;95(3):361–367. doi:10.1139/bcb-2016-011028177764
- KoppF, MendellJT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi:10.1016/j.cell.2018.01.01129373828
- Di StefanoA, CaramoriG, GnemmiI, et al. T helper type 17-related cytokine expression is increased in the bronchial mucosa of stable chronic obstructive pulmonary disease patients. Clin Exp Immunol. 2009;157(2):316–324. doi:10.1111/j.1365-2249.2009.03965.x19604272
- HuangP, YanR, ZhangX, WangL, KeX, QuY. Activating Wnt/beta-catenin signaling pathway for disease therapy: challenges and opportunities. Pharmacol Ther. 2019;196:79–90. doi:10.1016/j.pharmthera.2018.11.00830468742
- HoltzmanMJ, ByersDE, Alexander-BrettJ, WangX. The role of airway epithelial cells and innate immune cells in chronic respiratory disease. Nat Rev Immunol. 2014;14(10):686–698. doi:10.1038/nri373925234144
- ZhuZ, GongW, WangL, BaiY, YuZ, ZhangL. Efficient assessment of 3D train-track-bridge interaction combining multi-time-step method and moving track technique. Eng Struct. 2019;183:290–302. doi:10.1016/j.engstruct.2019.01.036
- ChenY, LuoH, KangN, et al. Beraprost sodium attenuates cigarette smoke extract-induced apoptosis in vascular endothelial cells. Mol Biol Rep. 2012;39(12):10447–10457. doi:10.1007/s11033-012-1924-123053949
- DabralS, TianX, KojonazarovB, et al. Notch1 signalling regulates endothelial proliferation and apoptosis in pulmonary arterial hypertension. Eur Respir J. 2016;48(4):1137–1149. doi:10.1183/13993003.00773-201527471204
- Artavanis-TsakonasS, RandMD, LakeRJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284(5415):770–776. doi:10.1126/science.284.5415.77010221902
