Abstract
Background
The detection of Pneumocystis jirovecii DNA in respiratory specimen from individuals who do not have signs or symptoms of pneumonia has been defined as colonization. The role of P. jirovecii colonization in the development or progression of various lung diseases has been reported, but little information about P. jirovecii colonization in patients is available in the People’s Republic of China.
Objective
To determine the prevalence of P. jirovecii colonization in patients with various pulmonary diseases, including the acute and stable stage of COPD, interstitial lung diseases, cystic fibrosis, and chronic bronchiectasis.
Materials and methods
A loop-mediated isothermal amplification (LAMP) and a conventional polymerase chain reaction (PCR) method for detecting P. jirovecii were developed. Ninety-eight HIV-negative patients who were followed-up and who had undergone bronchoscopy for diagnosis of various underlying respiratory diseases were included in the study. Sputa of these patients were analyzed with LAMP amplification of P. jirovecii gene. In addition, conventional PCR, Giemsa and Gomori’s methenamine silver nitrate staining assays were applied to all specimens.
Results
The sensitivity and specificity test showed that there was no cross-reaction with other fungi or bacteria in detecting the specific gene of P. jirovecii by LAMP, and the minimum detection limits by LAMP was 50 copies/mL. P. jirovecii DNA was detected in 62 of 98 (63.3%) sputa specimens by LAMP assay and 22.45% (22/98) by conventional PCR. However, no P. jirovecii cysts were found by Giemsa and Gomori’s methenamine silver nitrate in all of gene-positive specimens.
Conclusion
The results of our study showed that prevalence of P. jirovecii colonization is particularly high in patients with chronic pulmonary diseases in the People’s Republic of China, and the LAMP method is better for evaluation of the colonization of P. jirovecii in sputum specimen than conventional PCR.
Introduction
A high prevalence of Pneumocystis jirovecii colonization was observed in patients with chronic pulmonary diseases or various lung underlying diseases,Citation1–Citation3 and the important roles of P. jirovecii colonization in development or progression of various lung diseases is a concern.Citation4–Citation6 Patients who are carriers of P. jirovecii are at a higher risk of P. jirovecii pneumonia, and could also present a problem for public health since colonized patients could act as a major reservoir and source of infection for susceptible subjects.Citation7,Citation8 Detection of carriage or colonization of P. jirovecii is important for understanding its epidemiology and its correlation with the lung disease. Even though a number of Pneumocystis pneumonia (PCP) cases has been reported in the People’s Republic of China,Citation9 scarce data is available regarding the carriage or colonization of P. jirovecii in immunocompetent patients. Loop-mediated isothermal amplification (LAMP) is an innovative molecular technique to amplify a specific target gene.Citation10–Citation12 To evaluate the prevalence of P. jirovecii colonization in patients with pulmonary diseases, we have developed and evaluated a LAMP method to detect the P. jirovecii gene from sputum specimens of the patients with chronic pulmonary diseases. The specimens were also microbiologically examined.
Materials and methods
Patients and specimens
Ninety-eight HIV-negative patients suffering from chronic pulmonary diseases were incorporated in this study. They were consecutively treated in Department of Internal Medicine of the First Affiliated Hospital of China Medical University from June 2011 to October 2013. Every patient underwent a clinical and biochemical examination using a standardized protocol, and HIV antibody was tested using Anti-HIV−1+2 antibodies ELISA diagnostic kit. The patients had undergone bronchoscopy for diagnosis of various underlying respiratory diseases. The inclusion criteria was in hospital treatment for chronic pulmonary disease, including acute exacerbations of COPD (AECOPD), stable stage of COPD, interstitial lung diseases (ILDs), cystic fibrosis (CF), and chronic bronchiectasis (CB) patients. The diseases were diagnosed according to the People’s Republic of China Ministry of Health in 2011 to develop the disease diagnostic criteria.
Specimen processing
Blood and sputum specimens were collected from 98 patients before they received the corticoid or antibiotics treatment. Informed consent was obtained from all patients.
Sputum specimens were obtained from patient by spontaneous production. In order to avoid contamination of oral microbe, patients first gargled saline three times, prior to coughing up the sputum from deep respiratory tract when collecting specimens. Sputum specimens were promptly sent to the central laboratory of the hospital for bacterial culture and quantization. Co-infecting bacteria was analyzed using the automated bacterial identification system (ATB system, BioMérieux, Marcy-l’Étoile, France) following the bacterial or fungi culture. P. jirovecii organisms were identified microscopically by a modified Giemsa staining (Diff-Quik) method and a modified Gomori’s methenamine silver nitrate staining method as previously described.Citation13,Citation14
The blood samples were used for CD4+ T-cell measurement.Citation12 Sera were analyzed on BD FACSCalibur and the test kit. The normal critical value of CD4+ T-cell in blood was set to 410 cells/mm3 according to the reference of test kit. Patients will be considered as immunocompetent when CD4 cell >410 cells/mm3, and they will be identified as immunocompromised when the CD4+ T-cell counts <410 cells/mm3.
DNA preparation
DNA was extracted from the sputum specimens.Citation15 The sputa were treated with sodium hydroxide and centrifuged at 12,000 rpm for 5 minutes. The supernatant was discarded, and the precipitate was washed three times with TE buffer. Precipitated specimens were mixed with distilled water, incubated at 100°C for 15 minutes and centrifuged at 12,000 rpm for 10 minutes. The supernatants were stored at −20°C until DNA extraction. Following proteinase K digestion, DNA was extracted with a Genomic DNA Kit (Tiangen Technology, Beijing, People’s Republic of China) in accordance with the manufacturer’s instructions and stored at −80°C.
Design of primers
The gene sequences of Pneumocystis core ribosomal bodies small subunit 16s rRNA was obtained from Gen-Bank, and compared with the 16s rRNA of Pneumocystis carinii (AB266392), Pneumocystis murina (AY532651), Blastocystis, and Candida species. The specific LAMP primer was created by web tool Primer Explorer V4 (http://primerexplorer.jp/elamp4.0.0/index.html). Primers and their sequences used in LAMP are showed in . LAMP primers F3 and B3 were also used as conventional polymerase chain reaction (PCR) primers.
Table 1 Primers and their sequences used in LAMP
LAMP assay
The LAMP reaction was performedCitation11 in a final volume of 25 µL which contained 1× LAMP reaction buffer (the buffer contained 10 mM Tris-HCl (pH 8.8), 5 mM KCl, 5 mM (NH4)2SO4, 4 mM MgSO4, 0.1% Tween 20, and 0.4 M betaine (Sigma-Aldrich Co., St Louis, MO, USA)), 8 units of the Bst DNA polymerase (New England Biolabs, Ipswich, MA, USA), 100 ng of test DNA, and primers of 40 pmol (FIP and BIP), 20 pmol (Loop B and Loop F), and 5 pmol (F3 and B3). The mixture was incubated at 63°C in a water bath for 60 minutes. Detection of LAMP products was through visual fluorescence by adding 4× SYBR Green I (Thermo Fisher Scientific, Waltham, MA, USA) to 25 mL of LAMP product. The LAMP product was also electrophoresed on 1.5% agarose gel and visualized by UV gel imaging analyzer (Gel Imaging System, Shinova Co. Ltd., Shanghai, People’s Republic of China); or by observing the fluorescence collection volume by a real-time PCR instrument (ABI 7500; Thermo Fisher Scientific) for 60 minutes.
PCR and plasmid preparation
PCR was performed with the F3 and B3 primers as forward and reverse primers to confirm their specificity. A previously described PCR techniqueCitation14,Citation15 was used to amplify the Pneumocystis mitochondrial 16s rRNA gene. PCR conditions were as follows: the reaction was denatured at 95°C for 2.5 minutes, followed by 35 cycles of amplification at 94°C for 1 minute, 65°C for 1 minute, and 72°C for 1 minute, then with a final extension step at 72°C for 5 minutes. Finally, the PCR products were analyzed by electrophoresis and stained with ethidium bromide. The PCR product was ligated into the TA cloning vector pMD18-T (Takara BIO Co., Ltd., Dalian, People’s Republic of China). To prevent false-positives from contamination, pipettes with filters were used in all procedures. DNA extraction, preparation of the reaction mixture, PCR amplification, and detection were performed in separate areas under a laminar flow hood. To detect any cross-contamination, samples included a blank control or distilled water.
Sensitivity and specificity of the LAMP assay
The sensitivity of the LAMP method was examined using serial dilutions of Pneumocystis DNA copies in a plasmid solution. Detection limit of LAMP were measured and compared with that of the PCR. The specificity of the LAMP detecting was validated through the parallel and comparing test with Candida albicans, Candida tropicalis, Candida parapsilosis, and Candida glabrata, Mycoplasma pneumoniae (FH strain), Hemolytic streptococcus, methicillin-resistant Staphylococcus aureus strains in the same condition. Equal amounts of genomic DNA for each bacterium at 100 ng were used as the template. The specificity was also compared through detecting the sputum specimens of patient, the lungs of PCP positive model rat, or negative rat (ten rats, respectively). The amplified products of LAMP were examined separately using real-time turbidity, visualized by either the naked eye or using ultraviolet light after adding SYBR Green I, and tested by gel electrophoresis. The LAMP positive product was confirmed by sequencing with 16S rRNA of P. jirovecii.
Statistical analysis
Pearson chi-square test and continuous correction chi-square test were used for assessing differences between samples. Fisher’s exact test was used when the samples number <40 or the expected value <1. Results were considered significant at P>0.05. Statistical analyses were performed using the Statistical Package for Serial Studies for personal computers (SPSS version 12; SPSS Inc., Chicago, IL, USA).
Ethics
This study was conducted in accordance with the amended Declaration of Helsinki, and approved by the Institutional Review Board of Shengjing Hospital and the First Affiliated Hospital, China Medical University (AF-SOP-07-1). Informed consent was obtained from all patients.
Results
Ninety-eight patients were included in this study. The mean age of patients was 62±9.45 years (from 22 to 91 years old). Fifty-six (57.1%) were male. Distribution according to pulmonary diseases was 48.0% (47/98) COPD, including 15 acute stage and 32 stable stage of COPD; 25.5% (25/98) ILDs, including five idiopathic pulmonary fibrosis, five fibro-sing lung disease associated with connective tissue disease, three sarcoidosis, eight cryptogenic organizing pneumonia, and four diffuse alveolar hemorrhage; 18.4% (18/98) CF and 8.2% (8/98) CB ().
Table 2 Pneumocystis jirovecii colonization in patients with chronic lung diseases
The LAMP assay
The results of sensitivity tested for LAMP method compared with PCR was shown in . The minimum detection limits for LAMP was 50 copies/mL, whereas the conventional PCR could amplify 104 copies/mL. The specificity of the LAMP method showed that there was no cross-reaction with C. albicans, C. tropicalis, C. parapsilosis, and C. glabrata, M. pneumoniae (FH strain), H. streptococcus, methicillin-resistant S. aureus strains. The results of LAMP detecting the Pneumocystis gene in the sputum specimens of patient and the lungs of PCP model rats were positive, but it was negative in control rats. The positive results were consistent with the three examined methods, and difference of the positive and negative are apparent in .
Figure 1 Sensitivity of LAMP method compared with PCR.
Notes: Detection limit of LAMP or PCR assays were performed using serial tenfold dilutions of the Pneumocystis plasmid. (A) LAMP method: line 1, ×109 copies/mL; line 2, ×108 copies/mL; line 3, ×107 copies/mL; line 4, ×106 copies/mL; line 5, ×105 copies/mL; line 6, ×104 copies/mL; line 7, ×103 copies/mL; line 8, ×102 copies/mL; line 9, 50 copies/mL; line 10, 10 copies/mL. (B) PCR method: lane M, 100 bp DNA marker; lane 1, ×109 copies/mL; lane 2, ×108 copies/mL; lane 3, ×107 copies/mL; lane 4, ×106 copies/mL; lane 5, ×105 copies/mL; lane 6, ×104 copies/mL; lane 7, ×103 copies/mL; lane 8, ×102 copies/mL; lane 9, 50 copies/mL; lane 10, 10 copies/mL.
Abbreviations: LAMP, loop-mediated isothermal amplification; PCR, polymerase chain reaction.
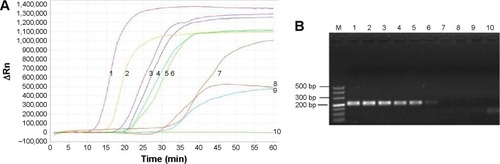
Figure 2 Examination of the products of LAMP for detecting the Pneumocystis jirovecii gene.
Abbreviations: LAMP, loop-mediated isothermal amplification; PCP, Pneumocystis pneumonia.
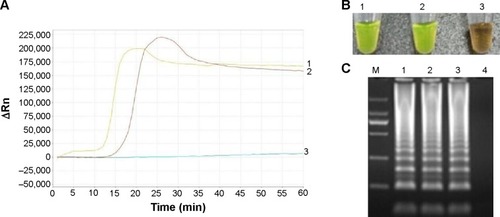
The gene were detected in 62 (63.3%) of the 98 patients by the LAMP method and 22/98 (22.5%) by conventional PCR (P<0.05). The positive rate of P. jirovecii was 53.2% in COPD patients, and it was 72.0%, 77.8%, and 62.5% in ILDs, CF, and CB patients, respectively (). No P. jirovecii organism was found in any of the 98 sputum specimens by Giemsa and Gomori’s methenamine silver nitrate stain. The results of co-infecting identification showed that co-infecting bacteria or fungus was detected in 62 cases, and the positive rate of P. jirovecii was 63.3% in co-infecting patients, and 75.0% in non-co-infecting patients, but was not statistically different (P>0.05). The species of microorganism in co-infecting patients were listed in , showing no statistical difference (P>0.05). The species of co-infecting microorganism in patients with various pulmonary disease subgroups were listed in , no significant difference was found.
Table 3 Microbes co-infection in 62 Pneumocystis jirovecii positive patients
Table 4 Microbe infection in the patients with difference pulmonary diseases subpopulation
The circulating CD4+ lymphocyte counts were tested by the flow cytometry. The detected results are shown in . The P. jirovecii positive rate in <410 cells/mm3 subjects was more than in CD4 counts >410 cells/mm3 subjects (80.0% vs 57.5%, P<0.05).
Table 5 Pneumocystis colonization in patients with different CD4+ T-cells
Discussion
As an innovative molecular technique, LAMP requires only a heating device and isothermal conditions to amplify a specific target gene. It is a highly efficient, selective, and rapid DNA amplification technique for genetic screening of pathogens and had developed for the diagnosis of PCP.Citation10–Citation12 In this study, we developed a LAMP method, compared with the conventional PCR method, and detected the P. jirovecii specific gene from sputum specimens of patients with chronic pulmonary diseases. The results showed that there was no cross-reaction with other fungi or several bacteria in detecting the specific gene of P. jirovecii by the LAMP method, and the minimum detection limits by the LAMP was 50 copies/mL, whereas the conventional PCR could amplify 104 copies/mL, it is at least 200 times more sensitive than PCR detection for sputum specimens. The reason for the difference between the two methods was that the LAMP uses three pairs of primers, which can be amplified by the loop primers, which can greatly improve the detection efficiency. The LAMP method developed by Japanese scholars was used for the diagnosis of PCP;Citation11,Citation12 we first applied LAMP for detection the P. jirovecii colonization in patients with chronic pulmonary diseases. The results prove that the LAMP method is an applicable technology for evaluating the colonization of P. jirovecii from sputum specimens. The method is rapid, simple, cost effective, and less time-consuming.
The P. jirovecii is an atypical opportunistic fungus that causes pneumonia in immunosuppressed and immunocompetent individuals. The presence of P. jirovecii DNA in respiratory specimens without clinical signs and symptoms of pneumonia has been defined as “colonization” or “carriage”, “subclinical infection” or “latent infection” P. jirovecii colonization in HIV-negative patients with various lung diseases has been reported.Citation2,Citation16–Citation19 The positive rate of P. jirovecii DNA is 33.7%Citation16 and 70%Citation17 in bronchoalveolar lavage specimens in Turkey; 17.3% in COPD patients in North LebanonCitation18 and 7.3% in Iranian patients with various lung diseases,Citation2 and 7.9% in COPD patientsCitation19 from pulmonary specimens. All of them used the nested PCR for detected gene of P. jirovecii. Here, we detected the colonization of P. jirovecii by LAMP and PCR method in sputa of HIV (−) patients with chronic pulmonary diseases. The results show that the positive rate of P. jirovecii DNA was up to 63.3% by LAMP method in sputa specimens of the patients with chronic pulmonary diseases in the People’s Republic of China. It is much higher than previous reports. The reason may be related to higher sensibility of LAMP (the minimum detection limits by the LAMP was 50 copies/mL) than nested PCR. The role of the higher yield of P. jirovecii colonization in the respiratory diseases remains to be determined. The carriers of P. jirovecii are at a higher risk of P. jirovecii pneumonia, or they may transmit the fungus to other susceptible patients.Citation7,Citation8 Preventive measures should be given more concern.
In the last few years, there is growing evidence that the lung microbe has a significant impact on clinical outcome of chronic respiratory diseases.Citation20–Citation22 The relationship of the lung microbes with the P. jirovecii colonization and the role in the lung disease had drawn attention.Citation23,Citation24 We tested the microbes in sputum and investigated whether the P. jirovecii colonization may inhibit or promote by other microorganism in lung. The results show that more than ten microbes were discovered in the sputum in 62 patients, but did not show any statistical difference between P. jirovecii colonization with co-infection and those without co-infection, and even co-infection with different bacterial species. However, we detected the microbes in the sputum specimens, which could not really show the true picture of microbial population in the lung, and it is with the risk of contamination of oral microbes, despite the preventive measures taken to avoid oral microbial contamination. It will be more reliable to test the lung microbes in bronchoalveolar lavage fluid specimens.
A series of animal and human studies suggest that the Pneumocystis infection is strongly correlated with CD4+ T-cell responses.Citation25 Pneumocystis colonization occurred when the CD4(+) T-cell count decreased to <500 cells/mL.Citation26 In this study, 80.0% of Pneumocystis gene was tested positive in the group of CD4+ T-cells counts <410/mm3, which was significantly higher than 57.5% in the CD4+ T-cells counts >410/mm3 group (P>0.05, ). These results support the findings of the importance of CD4+ T-cells in defense against Pneumocystis, the high prevalence of Pneumocystis colonization even in immunocompetent patients with high CD4+ T-cells counts.
The results of this study showed that prevalence of P. jirovecii colonization is particularly high in patients with chronic pulmonary diseases in the People’s Republic of China, suggesting that those patient population is a major reservoir and source of P. jirovecii infection and that identification of such individuals may contribute to PCP prevention strategies. The results of detection indicate that LAMP was a good method for evaluation of the colonization of P. jirovecii in sputum specimen.
Author contributions
Dong-Dong Wang developed the study design and carried out acquisition and interpretation of data, statistical analysis, manuscript preparation, and critical revision of intellectual contents. Dr Ming-Quan Zheng reviewed and edited the manuscripts. Nan Zhang and Chun-Li An contributed to screening patients, collecting sputum specimen, gene detection, acquisition and interpretation of data, revision of the article for important intellectual contents, and final approval of the version to be published. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NSFC: 81370189, 81302271). The authors wish to acknowledge the staff members of the Division of Pulmonary, Department of Internal Medicine, Shengjing Hospital, and the First Affiliated Hospital, China Medical University, for their contribution to this study. The authors thank the staff members of Department of Microbiology and Parasitology for their helpful assistance.
Disclosure
The authors report no conflicts of interest in this work.
References
- RieboldDEnohDOKingeTNPneumocystis jirovecii colonisation in HIV-positive and HIV-negative subjects in CameroonTrop Med Int Health201419664365524645978
- KhodadadiHMirhendiHMohebaliMKordbachehPZarrinfarHMakimuraKPneumocystis jirovecii colonization in non-HIV-infected patients based on nested-PCR detection in bronchoalveolar lavage samplesIran J Public Health201342329830523641407
- IzadiMJonaidi JafariNSadraeiJThe prevalence of Pneumo-cystis jirovecii in bronchoalveolar lavage specimens of lung transplant recipients examined by the nested PCRJundishapur J Microbiol2014712e1351825741434
- MorrisANorrisKAColonization by Pneumocystis jirovecii and its role in diseaseClin Microbiol Rev201225229731722491773
- Hernández-HernándezFFréalleECaneiroPProspective multi-center study of Pneumocystis jirovecii colonization among cystic fibrosis patients in FranceJ Clin Microbiol201250124107411023015669
- PederivaMAWissmannGFriazaVHigh prevalence of Pneumocystis jirovecii colonization in Brazilian cystic fibrosis patientsMed Mycol201250555656022206262
- Gits-MuselliMPeraldiMNde CastroNNew short tandem repeat-based molecular typing method for Pneumocystis jirovecii reveals intrahospital transmission between patients from different wardsPLoS One201510e012576325933203
- Le GalSPougnetLDamianiCPneumocystis jirovecii in the air surrounding patients with Pneumocystis pulmonary colonizationDiagn Microbiol Infect Dis20158213714225801779
- WangXLWeiWAnCLRetrospective study of Pneumocystis pneumonia over half a century in mainland ChinaJ Med Microbiol201160Pt 563163821273373
- DhamaKKarthikKChakrabortySLoop-mediated isothermal amplification of DNA (LAMP): a new diagnostic tool lights the world of diagnosis of animal and human pathogens: a reviewPak J Biol Sci201417215116624783797
- UemuraNMakimuraKOnozakiMDevelopment of a loop-mediated isothermal amplification method for diagnosing Pneumocystis pneumoniaJ Med Microbiol200857Pt 1505718065667
- NakashimaKAoshimaMOhkuniYHoshinoEHashimotoKOtsukaYLoop-mediated isothermal amplification method for diagnosing Pneumocystis pneumonia in HIV-uninfected immunocompromised patients with pulmonary infiltratesJ Infect Chemother2014201275776125187511
- AnCLGigliottiFHarmsenAGExposure of immunocompetent adult mice to Pneumocystis carinii f. sp. muris by cohousing: growth of P. carinii f. sp. muris and host immune responseInfect Immu200371420652070
- AnCLLiSJiangLMasanobuTDetection of the Pneumocystis carinii by PCR and organism staining methodJ China Med Univ19992812728 in Chinese
- WakefieldAECuiverLMillerRFHopkinJMDNA amplification on induced sputum samples for diagnosis of Pneumocystis carinii pneumoniaLancet1991337137813791674765
- ÖzkoçSBayram DelibaşSErbaycuAEErgüdenCAkısüÇInvestigation of Pneumocystis jirovecii colonization in patients with pulmonary diseasesTurkiye Parazitol Derg201438421421925732877
- ÖzkoçSBayram DelibaşSInvestigation of Pneumocystis jirovecii pneumonia and colonization in iatrogenically immunosuppressed and immunocompetent patientsMikrobiyol Bul201549222123026167822
- KhalifeSAliouatEMAliouat-DenisCMFirst data on Pneumocystis jirovecii colonization in patients with respiratory diseases in North LebanonNew Microbes New Infect20156111426042187
- SheikholeslamiMFSadraeiJFarniaPColonization of Pneumocystis jirovecii in chronic obstructive pulmonary disease (COPD) patients and the rate of Pneumocystis pneumonia in Iranian non-HIV(+) immunocompromised patientsIran J Microbiol20135441141725848514
- NguyenLDViscogliosiEDelhaesLThe lung mycobiome: an emerging field of the human respiratory microbiomeFront Microbiol201568925762987
- SethiSChronic obstructive pulmonary disease and infection. Disruption of the microbiome?Ann Am Thorac Soc201411Suppl 1S43S4724437405
- SzeMAHoggJCSinDDBacterial microbiome of lungs in COPDInt J Chron Obstruct Pulmon Dis2014922923824591822
- SivamSSciurbaFCLuchtLADistribution of Pneumocystis jirovecii in lungs from colonized COPD patientsDiagn Microbiol Infect Dis2011711242821851870
- KlingHMShipleyTWPatilSPRelationship of Pneumocystis jirovecii humoral immunity to prevention of colonization and chronic obstructive pulmonary disease in a primate model of HIV infectionInfect Immun201078104320433020660609
- FitzpatrickMETedrowJRHillenbrandMEPneumocystis jirovecii colonization is associated with enhanced Th1 inflammatory gene expression in lungs of humans with chronic obstructive pulmonary diseaseMicrobiol Immunol201458320221124438206
- KlingHMShipleyTWPatilSMorrisANorrisKAPneumocystis colonization in immunocompetent and simian immunodeficiency virus-infected cynomolgus macaquesJ Infect Dis20091991899619014344
