Abstract
Purpose
Dysfunction of endothelium is associated with multiple pathological vascular diseases. However, how to regulate reendothelialization after vascular injury is not well defined. This study aims to determine whether and how Paeonol controls reendothelialization following artery injury.
Methods
The endothelium of murine carotid artery was denuded by catheter guide wires injury. H&E staining and IF staining were performed to determine whether Paeonol is critical for reendothelialization. BRDU Incorporation Assay, Boyden Chamber Migration Assay, Tube Formation Assay, and Spheroid Sprouting Assay were used to investigate whether Paeonol is involved in regulating proliferation and migration of endothelial cells. The underlying mechanism of how Paeonol regulates reendothelialization was determined by Molecular docking simulation and CO-IP Assay.
Results
Paeonol treatment significantly inhibits neointima formation in carotid artery ligation model by promoting proliferation and migration of endothelial cells. Mechanistically, Paeonol enhances c-Myc expression, consequently interacts with VEGFR2 results in activating VEGF signaling pathway, and eventually promotes reendothelialization after vascular injury.
Conclusion
Our data demonstrated that Paeonol plays a critical role in regulating vascular reendothelialization, which may be therapeutically used for treatment of pathological vascular diseases.
Introduction
Maintenance of endothelium integrity and function plays a pivotal role during ontogeny of organisms. Endothelium lining inside of heart and vessels not only forms an interface between circulating blood, vascular, and lymph wall, but also forms a barrier between vascular and tissues. The integrity of endothelium plays a pivotal role in regulating communication between circulating blood and tissues.Citation1 Endothelial cells can synthesize and secrete multiple substances, which are crucial in regulating vascular contraction and relaxation, and synthesize and secrete a variety of enzymes, which are critical in controlling blood clotting and platelet adhesion.Citation2
Endothelial dysfunction can be induced by vascular injury, such as stent implantation. Preexisting vascular diseases, especially atherosclerosis,Citation3 hypertension,Citation4 diabetes,Citation5 and some other conditions, including cigarette smoking,Citation6 alcohol,Citation7 high fat diet,Citation8 and aging,Citation9 which can accelerate endothelial dysfunction. Dysfunction of endothelium is characterized by not only alternation of morphology and motility but also changes in endothelial behaviors, including an imbalance in the relative contribution of endothelium-derived relaxing and contracting factors,Citation10 disrupted vascular tone,Citation11 increased vascular permeability-induced inflammatory reactions,Citation3 which contributes to regulate development of thrombus formation,Citation12 atherosclerosis,Citation3 vascular restenosis,Citation13 arterial aneurysm,Citation14 and arterial stiffness.Citation15
The endothelium can be repaired by resident endothelial cells, circulating endothelial progenitor cells and marrow release of endogenous progenitor cells, which mediates homing and arrangement in arterial denuded sites or intravascular stent surfaces.Citation16,Citation17 Endothelial cell-specific growth factor, including VEGF, promotes reendothelialization.Citation18 Studies have demonstrated that Neurotrophin-3 promotes reendothelialization by promoting endothelial progenitor cell mobilization and homing.Citation19 Pattern recognition receptors (PRRs) promote reendothelialization through increased inflammasome activation.Citation20 Mitochondrial oxidation caused by SIRT3/SOD2 signaling deficiency contributes to decline of reendothelialization.Citation21 Insulin and Bone marrow-derived Krüppel-like factor 10 accelerates reendothelialization following arterial endothelial injury.Citation22,Citation23 Erythropoietin-mobilized endothelial progenitors induce reendothelialization via Akt-mediated nitric oxide synthase activation.Citation24 Eva-1 homologue 1 (Eva1a) ameliorates atherosclerosis by promoting reendothelialization via Rac1 signaling pathway.Citation25
Numerous studies have demonstrated that Traditional Chinese Medicine could be used for cardiovascular disease treatment. Paeonol is a natural ingredient isolated from Paeonia Suffruticosa and Paeonol lactiflora pall.Citation26 Our previous studies have demonstrated that Paeonol suppresses SMC phenotypic switching associated with neointima formation by inhibiting SMC proliferation and mobility. However, whether inhibition of neointima formation is due to regulating reendothelialization is unknown. Our previous study also indicates that Paeonol treatment dramatically induces endothelial cell c-Myc expression. c-Myc is an early molecular marker of cell cycle activity.Citation27 Research has found that c-Myc could directly promote SMC growth and motilityCitation28,Citation29 and suppress vascular inflammation,Citation30 which is critical for the development of occlusive vascular disease.Citation27 However, whether c-Myc regulates reendothelialization is not well defined. This study aimed to investigate the potential role of Paeonol in regulating reendothelialization.
Materials and Methods
Ethics Approval and Consent to Participate
Animal care and treatment refer to the National Institutes of Health “Guide for the Care and Experimentation of Laboratory Animals”. The experimental protocols and operating procedures were reviewed and approved by the Animal Welfare and Use Committee of Chengdu University of Traditional Chinese Medicine (ethical approval number: 2019–04).
Chemicals
Paeonol is an effective ingredient of Paeonia suffruticosa Andr. Paeonol (C9H10O3) was purchased from Sigma. Ki8751 (C24H18F3N3O4) was purchased from Selleck.
Animal Model
Murine carotid artery catheter guide wires injury performed as in a previous publication.Citation31 Briefly, 2-month-old male ApoE−/− mice were pretreated with Paeonol (30 mg/kg) by intraperitoneal injection for 7 days. Mice were anesthetized and placed on a temperature-controlled heating pad. Separated internal carotid artery and external artery under microscope. Endothelium of common carotid artery was denuded by catheter guide wires (0.014 inch). Mice were continuously treated with Paeonol until sacrificed. The carotid arteries were harvested and paraffin-embedded for further analysis. The mice were anesthetized by intraperitoneal injection of ketamine 80 mg/kg and xylazine 5 mg/kg.
Hematoxylin and Eosin (H&E), Immunohistochemistry (IHC), and Immunofluorescence (IF) Staining
Harvested murine common carotid arteries, paraffin embedded, and collected slides of 5 μm thickness. H&E staining was performed as in our previous study.Citation32 For IHC staining, the slides were dealt with unmasking solution for antigen retrieval, followed by incubation with primary antibody. After being incubated with a biotinylated secondary antibody, ABC and DAB solutions (Vector Laboratories) were used to visualize targets. For IF staining, the slides were immersed in citric acid solution for antigen retrieval. Blocked and incubated with primary antibody. A fluorophore-conjugated secondary antibody was used. Images were captured using confocal microscopy (LS510, Zeiss).
Cell Culture
Human primary umbilical vein endothelial cell (HUVEC) and human umbilical vein/vascular endothelial cell line (HUVEC-C) were purchased from ATCC (USA). HUVEC cells were cultured in vascular cell basal medium (ATCC, USA) and supplemented with endothelial Cell Growth Kit (ATCC, USA), and the HUVEC-C cells cultured in basal medium (ATCC, USA) containing 10% FBS (Gibco, USA).
CCK-8 Assay
HUVECs were trypsinized and seeded in a 96-well culture plate (3×103/well), following Paeonol (10 μM) treatment for 24 h. After incubated of CCK-8 solution, the absorbance at 450 nm was measured.
BRDU Incorporation Assay
Suspended HUVEC in the medium containing Paeonol (10 μM), then labeled with BRDU labeling reagent for 12 h. IF staining was performed to visualize BRDU-incorporated cells.
Cell Cycle Analysis
Treated HUVEC with Paeonol (10 μM), trypsinized, and fixed with 70% ethanol. Cell cycle was analysed by flow cytometry analysis after propidium iodide staining (BD Biosciences).
Boyden Chamber Migration Assay
Cells seeded in serum-free medium that was settled into a culture plate containing 10% FBS and Paeonol (10 μM). Migrated cells were counted after crystal staining.
Spheroid Sprouting Assay
More details of spheroid sprouting assay described in our previous publication.Citation33 Mixed cell suspension and methylcellulose solution in a ratio of 1:4. The mixture was added in a 96-well culture plate to form “cell sphere”. Neutralized liquid collagen solution and solidified at 37°C. Calcein staining was used to visualize cells. Images of “cell spheres” were captured under microscope (Leica Microsystem CMS GmbH). Image J was used to quantify the length of spheroid sprouting.
Matrigel-Based Tube Formation Assay
Solidified neutralized collagen solution and suspended HUVEC-Cs with medium contained Paeonol (10 μM). The endothelial tubule formation was monitored under microscope and images were captured after Calcein staining. Cumulative tube numbers were quantified using Image J.
Quantitative Real Time PCR Analysis
Total RNA was extracted from HUVEC or HUVEC-C using the Trizol reagent (Thermo Invitrogen). Reverse transcription was performed using iScript cDNA synthesis kit (Vazyme). Quantification was calculated using the 2−ΔΔCT method against β-actin. The primer sequences are exhibited in primer list (Supplementary Figure 1).
Western Blot Analysis
Proteins from HUVEC or HUVEC-C were extracted using RIPA lysis buffer. Protein concentration was evaluated using BCA kit (Biosharp). Protein is denatured at 98°C for 10 min and separated by SDS-PAGE. It is incubated with specific antibodies. Visualization was performed using a chemiluminescence Western blotting detection kit, and images were captured by a chemiluminescence detection system.
Protein and Protein Interaction (PPI) Network Analysis
The String (Version: 11.5) database (http://string-db.org/) was used to obtain the protein–protein interactions (PPI). The organism was selected as Homo sapiens, interaction score ≥0.4 was set, and disconnected nodes were hidden in the network.
Molecular Docking
The molecular docking performed by Autodock Vina 1.5.6 was used to evaluate the binding energy. The mol2 format for the structure of Paeonol was downloaded through TCMSP (https://old.tcmsp-e.com/). The three-dimensional structures of c-Myc (Homo, ID: 5G1X) and VEGFR2 (Homo, ID: 1VR2) were downloaded from PDB database (http://www.rcsb.org/). Before molecular docking, we removed the organic and H2O molecules of the macromolecular protein and added hydrogens before molecular docking. Then, the Grid Box was prepared. Finally, the value of binding energy was measured.
Co-Immunoprecipitation Assay
Cell lysate is extracted from HUVEC cells. Cleared Protein A/G agarose beads (TransGen Biotech, Beijing, China). Mixed beads and cell lysate and incubated with c-Myc (Abcam) and VEGFR2 (Cell Signaling Technology). The CO-immunoprecipitation protein targets were evaluated using Western blotting.
Protein Docking Prediction
The protein docking prediction is based on a previous publication.Citation34 Briefly, the crystal structures of c-Myc (ID: 5G1X) and VEGFR2 (ID: 1VR2) were obtained from the RSCB PDB and the IDs of both were input into ZDOCK SERVER (http://zdock.umassmed.edu/), in which we obtained the complexes. Then, the complexes ranked top five were analyzed by PDBePISA (https://www.ebi.ac.uk/msd-srv/prot_int/). Finally, the complex ranked top was visualized by Pymol Version 2.5.2.
Statistical Analysis
Quantitative data was presented as mean ± SEM. GraphPad prism software was used for statistical analysis. The difference in significance between two groups was analyzed using unpaired Student’s t-test. One-way ANOVA followed by Tukey’s-b test and repeated measures ANOVA followed by S-N-K (Student-Newman-Keuls) test were used to compare the differences in cell viabilities after different doses of paeonol treatment and at different time points, respectively. Normal Q–Q plots are used to assess data distribution. * P < 0.05 was considered statistically significant.
Results
Paeonol Alleviates Vascular Stenosis Induced by Endothelial Injury
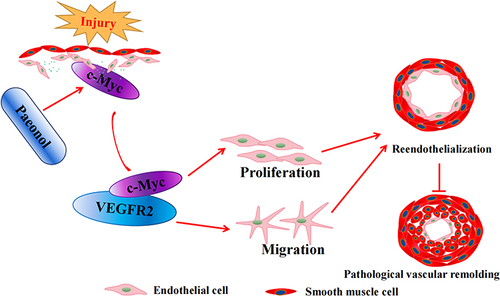
Various vascular diseases lead to vascular stenosis after endothelial injury. To assess whether Paeonol involves in regulating vascular stenosis responding to vascular injury, catheter guide wires injury was performed. The male ApoE−/− mice were pretreated with Paeonol (30 mg/kg) for consecutive 7 days by intraperitoneal injection. After catheter guide wires injury, those animals were continuously treated with Paeonol until tissues were harvested (). H&E was performed to observe vascular pathological changes. At the third day after surgery, most endothelial cells were denuded by catheter guide wires. Paeonol treatment did not change the thickness of the vascular wall, as well as lumen areas (Supplementary Figure 2A and B). However, at the seventh day after catheter guide wires, Paeonol treatment significantly suppressed vascular neointima formation characterized by decreased neointima areas, media layer areas, and ratio of neointima and media layer areas (). Similar results were observed at the 10th day after injury, when Paeonol treatment dramatically suppressed neointima areas, media layer areas, and the ratio of neointima and media layer areas (). Our IF staining against the CD31 antibody further indicated that a higher expression level of CD31 was exhibited around the inner neointima after Paeonol treatment (; Supplementary Figure 3). Our data showed that Paeonol attenuates vascular stenosis, possibly through vascular reendothelialization.
Figure 1 Paeonol attenuates vascular stenosis induced by vascular injury. (A) Schematic diagram of murine carotid artery catheter guide wires and Paeonol treatment. (B) Representative images of left carotid artery catheter guide wires injury after 7 days of Paeonol treatment of H&E staining (Scale bar: 50 μm). (C and D) Quantification of neointima area of left carotid artery cross-section and the ratio of neointima area to media area were analyzed by Image J (n=9). (E) Representative images of the left carotid artery after Paeonol treatment for 10 days of H&E staining (Scale bar: 50 μm). (F and G) Quantification of neointima area of left carotid artery cross-section and the ratio of neointima area to media area (n=9). (H) IF staining against CD31 and SM α-actin antibodies on the injured artery slides after Paeonol treatment for 7days (Scale bar: 100 μm). Quantitative data presented as mean ± SEM, *P < 0.05 was considered significant.
Paeonol Promotes Vascular Endothelial Cell Proliferation
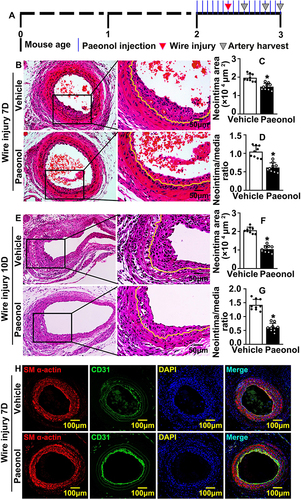
HUVEC was treated with Paeonol (10 μM) for 24 h, and our CCK-8 assay results indicated that Paeonol treatment enhanced HUVEC viability (). The number of HUVEC-Cs was counted, and we observed that Paeonol treatment significantly increased cell numbers following 24 h, 48 h and 72-h treatment (). Paeonol treatment regulated the transcription level of cell cycle-related genes characterized by suppressing cell cycle negative related genes as P14arf, P18ink4c, and P19arf, whereas increasing cell cycle positive related genes CCND1, which was determined by Real-Time PCR (). Based on the flow cytometry analysis after PI staining, we found that Paeonol treatment induced HUVEC-C to enter the synthesis phase during cell cycle (; Supplementary Figure 4). BRDU assay showed that Paeonol significantly increased the percentage of BRDU-positive HUVEC cells ( and ). We next sought to determine whether Paeonol induced endothelial cell proliferation in our animal studies. At the third day after vascular injury, the endothelial was undetectable due to catheter guide wires injury. However, at the seventh day after catheter guide wires injury, the endothelial cells mainly exhibited around the inner part of neointima and more proliferating endothelial cells displayed after Paeonol treatment, which was determined by using IF staining against CD31 and PCNA antibodies (). Similar results were also exhibited at the 10th day after vascular injury. Within the endothelial layer of neointima, Paeonol treatment induced PCNA and Ki67 expression, which was evaluated using IHC staining (Supplementary Figure 5). These data indicated that Paeonol induced endothelial cell proliferation response to vascular injury.
Figure 2 Paeonol promotes vascular endothelial cell proliferation. (A) CCK-8 Assay detecting HUVEC viability and proliferation after Paeonol (10 μM) treatment for 24 hours (n=8). (B) Cell numbers at different time points (24 hours, 48 hours, 72 hours) after HUVEC-Cs treated with Paeonol (n=6). (C) Real Time PCR detected mRNA expression levels of cell cycle-related genes (n=6). (D) HUVEC-Cs were treated with Paeonol for 48 hours. Cell cycle analyzed using flow cytometry analysis after Propidium Iodide (PI) staining (n=5). (E) BRDU incorporation assay performed to evaluate incorporation of BRDU into HUVECs (Scale bar: 20 μm). (F). Quantification of BRDU positive cells (n=6). (G) IF staining against CD31 and PCNA antibodies on slides after Paeonol treatment for 7 days (Scale bar: 100 μm. White triangles represented the positive signal of specific antibodies). Quantitative data presented as mean ± SEM, *P < 0.05 was considered significant.
Paeonol Promotes Vascular Endothelial Cell Motility
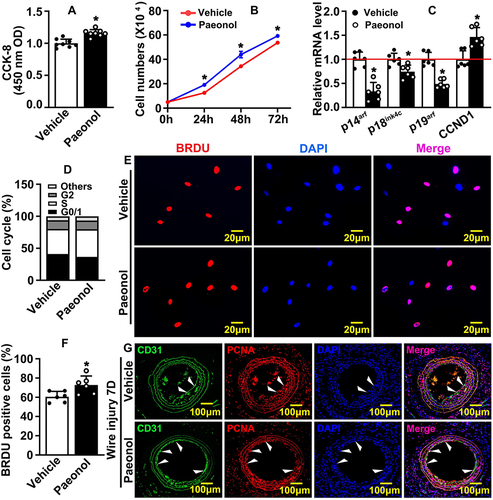
We performed Boyden migration assay and observed that Paeonol significantly induced HUVEC migration, and the number of HUVECs that passed through Boyden’s chamber was dramatically increased ( and ). Matrigel-based Tube Formation Assay was performed to evaluate whether Paeonol promotes endothelial cell regulated tube formation. After Paeonol treatment, more tube-like phenotypes, including tube number and partially tube-like branches, were exhibited comparing vehicle groups ( and ). We also performed Spheroid Sprouting Assay, which turned out that Paeonol observably enhanced both the number of sprouts and sprouts lengths (). The metabolism of extracellular matrix plays a pivotal role in regulating cell motility. After Paeonol treatment, we evaluated transcription levels of extracellular matrix-related genes using Real-Time PCR, and the results indicated that transcription levels of Collagen 1, Fibronectin, VE-Cadherin, and MMP9 dramatically increased (). Protein level of VE-Cadherin was also evaluated, and we observed that Paeonol observably induced VE-Cadherin expression in HUVEC( and ). Our data indicated that Paeonol can significantly induce the motility of endothelial cells.
Figure 3 Paeonol promotes vascular endothelial cell motility. (A) Boyden migration assay evaluating cell motility after HUVECs were treated with Paeonol. Images were captured after crystal violet staining (Scale bar: 50 μm). (B) Quantification of migrated cells (n=6). (C) Matrigel based tube formation assay was performed after HUVEC-C treated with Paeonol (Scale bar: 50 μm). (D) Quantification of tube numbers (n=6). (E) The migrating ability of HUVECs after Paeonol treatment was evaluated by spheroid assay after Calcein AM staining (Scale bar: 20 μm). Sprouting numbers and sprouting lengths were quantified using the Image J software and exhibited in (F and G) (n=6). (H) The mRNA levels of migration-related genes (n=6). (I) The expression of VE-Cadherin in HUVEC was determined by IF staining (Scale bar: 20 μm). (J) Fluorescence intensity (FI) of VE-Cadherin quantified (n=6). Quantitative data presented as mean ± SEM, *P < 0.05 was considered significant.
Paeonol Induces Endothelial Cell c-Myc Expression After Vascular Injury
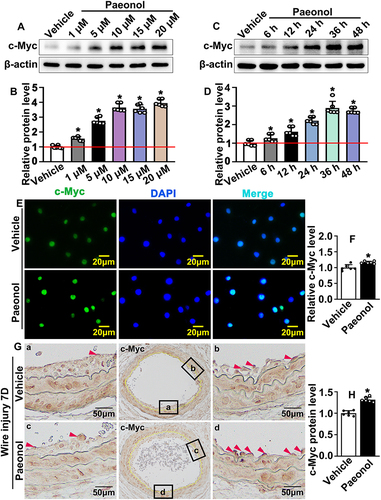
Based on our previous studies of Paeonol regulated pathological vascular remodeling by monitoring vascular smooth muscle cell phenotypic switching,Citation26 we hypothesized that Paeonol may contribute to reendothelialization, which is involved in pathological vascular remodeling. We treated HUVEC with Paeonol and performed Real-Time PCR to evaluate vasculogenic signaling pathways. Our results indicated that Paeonol could observably induce c-Myc, as well as VEGF signaling pathway expression (Supplementary Figure 6). We first sought to determine whether Paeonol directly regulates the expression level of c-Myc in endothelial cells. Paeonol treatment promoted c-Myc transcription levels in both HUVEC-C and HUVEC (Supplementary Figure 7A). We treated HUVEC-C with different doses of Paeonol, including 1 μM, 5 μM, 10 μM, 15 μM, and 20 μM. Proteins were harvested, and Western blot was performed. Our results indicated that 5 μM, 10 μM, 15 μM, and 20 μM Paeonol treatment observably induced c-Myc expression in HUVEC-C ( and ). After 10 μM of Paeonol treatment, c-Myc protein levels in HUVEC-C were evaluated at different time points, including 6 h, 12 h, 24 h, 36 h, and 48 h. Our Western blot data indicated that Paeonol treatment for 6 h significantly induced c-Myc expression in HUVEC-C, and a high expression of c-Myc could be detected after 48-h treatment ( and ). The expression of c-Myc in HUVEC-C also increased following Paeonol treatment (Supplementary Figure 7B). We further determined the expression pattern of c-Myc and observed that c-Myc was specifically expressed in nucleus, which can be enhanced following Paeonol treatment ( and ). We next sought to determine whether Paeonol induced c-Myc expression in our animal studies. At the third day after the catheter guide wires injury, lower expression of c-Myc was exhibited in the media smooth muscle layer, as well as in adventitia layer. Whereas no distinguishable difference was displayed between the Paeonol group and the vehicle group (Supplementary Figure 7C). At the seventh day after catheter guide wires injury, higher expression of c-Myc was exhibited within the media smooth muscle cell layer, neointima layer, and inner endothelial monolayer. Endothelial c-Myc dramatically increased after Paeonol treatment ( and ). Our data indicated that Paeonol induces endothelial cell c-Myc expression after vascular injury.
Figure 4 Paeonol induces endothelial cell c-Myc expression after vascular injury. (A) Western blot analysis of c-Myc protein expression in HUVEC-Cs after treated with different dose of Paeonol (1 μM, 5 μM, 10 μM, 15 μM, 20 μM). (B) Quantification of relative c-Myc protein after different dose of Paeonol treatment (n=6). (C) Western blot analysis of c-Myc protein expression in HUVEC-Cs after treated with Paeonol (10 μM) for different time points (6 hours, 12 hours, 24 hours, 36 hours, 48 hours). (D) Quantification relative c-Myc protein after different time points of treatment (n=6). (E) IF staining against c-Myc antibody performed in HUVEC-Cs after Paeonol treatment (Scale bar: 20 μm). (F) Fluorescence intensity of c-Myc was analyzed (n=6). (G) IHC staining against c-Myc antibody performed on slides after Paeonol treatment for 7 days (Scale bar: 50 μm. Red triangles represented the positive signal of specific antibodies). (H) c-Myc positive cells were analyzed (n=6). Quantitative data presented as mean ± SEM, *P < 0.05 was considered significant.
Paeonol Actives Endothelial Cell VEGF Signaling Pathway After Vascular Injury
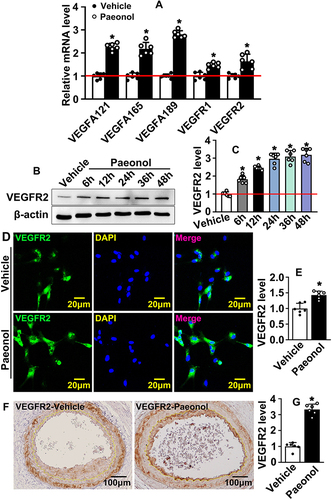
To confirm whether VEGF signaling pathway is involved in reendothelialization, we performed Real-Time PCR to evaluate targets of VEGF signaling pathway. Our results indicated that Paeonol treatment observably induced VEGFA121, VEGFA165, VEGFA189, VEGFR1, and VEGFR2 expression (). After Paeonol treatment, protein was harvested after 6 h, 12 h, 24 h, 36 h, and 48 h. Our Western blot results indicated that Paeonol treatment dramatically induced VEGFR2 expression after 6 h, and high expression levels of VEGFR2 were detectable from 6 h to 48 h ( and ). VEGFR2 ubiquitin is expressed in HUVEC and could be significantly enhanced after Paeonol treatment ( and ). We further evaluated whether Paeonol treatment induced VEGFR2 expression in our animal studies. For catheter guide wires injury model, Paeonol treatment dramatically induced VEGFR2 expression, and high expression of VEGFR2 was exhibited in the inner layer of neointima, which was determined by IHC staining ( and ). The skeletal injury induced angiogenesis model was also used in our studies. Our IF staining against CD31 and VEGFR2 antibodies indicated that Paeonol could induce endothelial cell VEGFR2 expression during skeletal muscle injury induced angiogenesis (Supplementary Figure 8). Our data indicated that Paeonol activates endothelial cell VEGF signaling pathway after vascular injury.
Figure 5 Paeonol actives endothelial cell VEGF signaling pathway after vascular injury. (A) The mRNA levels of VEGF signaling pathway-related genes in HUVECs after Paeonol treatment (n=6). (B) The protein level of VEGFR2 protein in HUVEC-Cs treated with Paeonol for different time points. (C) The quantification relative protein of VEGFR2 (n=6). (D) IF staining against VEGFR2 antibody performed to evaluate VEGFR2 expression in HUVEC after Paeonol treatment (Scale bar: 20 μm). (E) The quantification of the relative fluorescence intensity of VEGFR2 after Paeonol treatment (n=6). (F) IHC staining against VEGFR2 antibody performed to detect VEGFR2 expression on slides after Paeonol treatment for 7 days (Scale bar: 100 μm). (G) Integrated optical intensity analyzed (n=6). Quantitative data presented as mean ± SEM, *P < 0.05 was considered significant.
Paeonol Induces Reendothelialization by Regulating the Interaction Between c-Myc and VEGFR2 After Vascular Injury
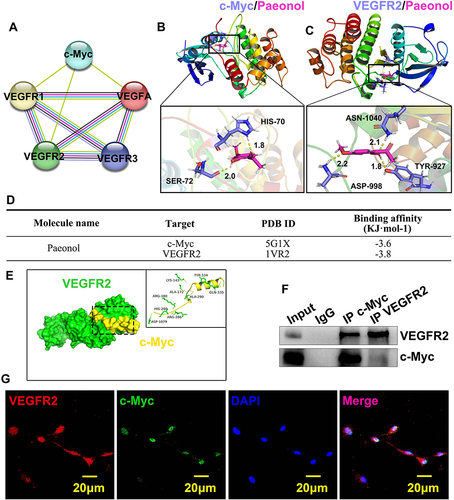
Both c-Myc and VEGFR2 contributed to reendothelialization. However, whether c-Myc interacts with VEGFR2 is largely unknown. We first analyzed protein–protein interaction using the STRING database (https://www.string-db.org/), including c-Myc, VEGFA, VEGFR1, VEGFR2, and VEGFR3. Our analysis of data suggested that c-Myc can interact with VEGFA, VEGFR1, and VEGFR2 (). To next seek to evaluate the interaction between Paeonol and c-Myc, VEGFR2, we conducted Molecular docking and measured the binding energy of Paeonol with c-Myc and VEGFR2. Multiple potential binding sites within c-Myc and VEGFR2 have been displayed ( and ). When the value of binding energy is less than zero, those binding sites are considered spontaneously binding and interacting with paeonol. −3.6 of binding energy exhibited between Paeonol and c-Myc, and −3.8 of binding energy exhibited between Paeonol and VEGFR2 (). We further tried to predict the potential binding sites between c-Myc and VEGFR2 using ZDOCK (). 3D structures of c-Myc and VEGFR2 were obtained from RCSB database (https://www.rcsb.org). c-Myc served as ligand and VEGFR2 served as acceptor. The potential binding sites identified using ZDOCK database (https://zdock.umassmed.edu/). The potential binding sites between c-Myc and VEGFR2 can form a complex that was determined by binding energy and interface area using PDBePISA database (https://www.ebi.ac.uk/msd-srv/prot_int/). Our data indicated that five complexes exhibited which likely the potential binding sites between c-Myc and VEGFR2 (Supplementary Figure 9). We also performed a CO-immunoprecipitation Assay to evaluate the interaction between c-Myc and VEGFR2. Total proteins prepared from HUVEC, and after antibodies immunoprecipitation, we observed that c-Myc could bind to VEGFR2 (). We next sought to determine expression patterns c-Myc and VEGFR2 in HUVEC. Our IF staining against c-Myc and VEGFR2 antibodies indicated that c-Myc and VEGFR2 CO-expressed in the nucleus (). Those data demonstrated that endothelial cell c-Myc interacts with VEGFR2 to promote reendothelialization after vascular injury.
Figure 6 Paeonol induces reendothelialization by regulating interaction between c-Myc and VEGFR2 after vascular injury. (A) The interactions among c-Myc, VEGFA and VEGFR family were analysed by String Database. Nodes represent proteins while edges represent interaction. (B and C) Molecular docking simulation performed to evaluate interaction between Paeonol with c-Myc protein and VEGFR2 protein. (D) Binding affinity of Paeonol to protein c-Myc and VEGFR2 was analyzed. (E) The complex of VEGFR2 and c-Myc docking processed by ZDOCK. VEGFR2 is shown in green, c-Myc in red. (F) CO-IP was performed to evaluate the interaction between c-Myc and VEGFR2. (G) IF staining performed in HUVECs to exhibit co-localization of c-Myc and VEGFR2 (Scale bar: 20 μm).
Inhibition of the VEGF Signaling Pathway Attenuates Paeonol Associated Reendothelialization, Which is Essential in Suppressing Pathological Vascular Remodeling
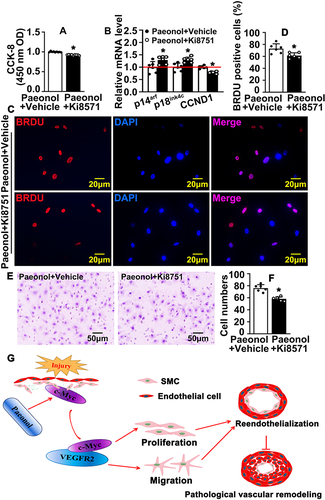
To identify whether Paeonol regulates reendothelialization specifically through c-Myc/VEGF Pathway, we inhibited VEGF signaling pathway by Ki8751 treatment, a VEGFR2 inhibitor, to suppress VEGFR2. Paeonol treatment promoted HUVEC-C viability, whereas VEGFR2 inhibition can attenuate Paeonol treatment induced HUVEC-C viability (). After inhibition of the VEGF signaling pathway following Paeonol treatment, we used Real-Time PCR to evaluate cell cycle-related genes and observed that transcription levels of P14arf, P18ink4c were reduced, whereas CCND1 increased (). After inhibition of the VEGF signaling pathway following Paeonol treatment, BRDU incorporation assay data indicated that inhibition of the VEGF signaling pathway could attenuate Paeonol treatment induced BRDU-positive cells ( and ). Inhibition of the VEGF signaling pathway can also reduce Paeonol treatment induced cell motility ( and ).
Figure 7 Inhibition of VEGF signaling pathway attenuates Paeonol associated reendothelialization. (A) HUVEC-Cs were treated with 10 μM Paeonol and 10 nM Ki8571 (VEGFR2 inhibitor), and CCK-8 performed to evaluate cell viability (n=9). (B) HUVEC-Cs were treated with Ki8751 (10 nM) after treatment with Paeonol. Real Time PCR performed to detect the relative mRNA levels of cell cycle-regulated genes (n=6). (C) HUVEC-Cs were treated with Ki8571 (10 nM) and Paeonol (10 μM) for 12 hours, after BRDU reagent labelled for 24 hours (Scale bar: 20 μm). (D) BRDU positive cells were quantified (n=6). (E) Boyden migration assay was performed after Paeonol (10 μM) and Ki8571 (10 nM) treating for 24 hours, and cell numbers were quantified in (F) (Scale bar: 50 μm; n=6). Quantitative data presented as mean ± SEM, *P < 0.05 was considered significant. (G) Schematic diagram demonstrates that how Paeonol regulates reendothelialization after vascular injury.
Discussion
This study provides evidence that Paeonol promotes reendothelialization following artery injury. Paeonol induces c-Myc expression, which is associated with VEGFR2, to accelerate reendothelialization and eventually suppress pathological vascular remodeling ().
Vascular endothelial cell exhibits high plasticity.Citation35,Citation36 High plasticity is critical not only for vascular development but also important for endothelium repair. Repair of endothelium is mediated by proliferation, differentiation and arrangement of resident endothelial cells, circulating endothelial progenitor cells and marrow release of endogenous progenitor cells.Citation16,Citation17 In this study, we demonstrated that Paeonol could induce reendothelialization after endothelial denuded by catheter guide wires injury, and our in vitro studies on HUVEC also indicated that Paeonol can promote cell proliferation and motility. However, whether existent resident endothelial cells or circulating endothelial progenitor cells and marrow release of endogenous progenitor cells are regulated by Paeonol is not clearly elucidated. Further studies will focus on identifying the major cell source that controls reendothelialization after artery endothelial injury.
Some other cell types, including SMC, fibroblast, and adipocyte, contribute to reendothelialization after artery injury. Our previous study demonstrated that Paeonol suppressed pathological vascular remodeling by regulating SMC phenotypic switching.Citation26 Dysfunction of endothelium leads to enhanced vascular permeability-induced vascular inflammatory response. However, it is hard to convince the relationship between inflammatory stress and pathological vascular remodeling due to the different stages of vascular diseases. More studies combined with clinical observation are required to investigate the function of crosstalk between endothelial cells and SMCs.
Normal primary human endothelial cells (HUVEC) and human endothelial cell line (HUVEC-C) were used in this study. However, our data indicated that HUVEC is more sensitive in responding to Paeonol treatment than HUVEC-C. Different doses of Paeonol treatment dramatically increased the viability of HUVEC, whereas no difference was exhibited in HUVEC-C after Paeonol treatment (Supplementary Figure 10A–D). Both vascular catheter guide wire injury and skeletal muscle injury models were used. Difference in morphology, functions, and behaviors of endothelial cells was exhibited between arteries and developing endothelial cells.
Conclusion
In summary, our data suggested that Paeonol could significantly accelerate reendothelialization after vascular injury through activating c-Myc-mediated VEGFR2 signaling pathway at least partially, which provides a potential strategy for endothelial repair.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval for the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
Additional information
Funding
References
- Escribano J, Chen MB, Moeendarbary E, et al. Balance of mechanical forces drives endothelial gap formation and may facilitate cancer and immune-cell extravasation. PLoS Comput Biol. 2019;15(5):e1006395. doi:10.1371/journal.pcbi.1006395
- Vanhoutte PM, Shimokawa H, Feletou M, Tang EH. Endothelial dysfunction and vascular disease - a 30th anniversary update. Acta physiologica. 2017;219(1):22–96. doi:10.1111/apha.12646
- Gimbrone MA Jr., García-Cardeña G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res. 2016;118(4):620–636. doi:10.1161/CIRCRESAHA.115.306301
- Spieker LE, Flammer AJ, Lüscher TF. The vascular endothelium in hypertension. Handb Exp Pharmacol. 2006;2006(176 Pt 2):249–283.
- Karan A, Bhakkiyalakshmi E, Jayasuriya R, Sarada DVL, Ramkumar KM. The pivotal role of nuclear factor erythroid 2-related factor 2 in diabetes-induced endothelial dysfunction. Pharmacol Res. 2020;153:104601. doi:10.1016/j.phrs.2019.104601
- Messner B, Bernhard D. Smoking and cardiovascular disease: mechanisms of endothelial dysfunction and early atherogenesis. Arterioscler Thromb Vasc Biol. 2014;34(3):509–515. doi:10.1161/ATVBAHA.113.300156
- Naik VD, Davis-Anderson K, Subramanian K, Lunde-Young R, Nemec MJ, Ramadoss J. Mechanisms underlying chronic binge alcohol exposure-induced uterine artery dysfunction in pregnant rat. Alcohol Clin Exp Res. 2018;42(4):682–690. doi:10.1111/acer.13602
- Cao Q, Xu D, Chen Y, et al. Sitagliptin reduces endothelial dysfunction and apoptosis induced by high-fat diet and palmitate in thoracic aortas and endothelial cells via ROS-ER stress-CHOP pathway. Front Pharmacol. 2021;12:670389. doi:10.3389/fphar.2021.670389
- Donato AJ, Morgan RG, Walker AE, Lesniewski LA. Cellular and molecular biology of aging endothelial cells. J Mol Cell Cardiol. 2015;89(Pt B):122–135. doi:10.1016/j.yjmcc.2015.01.021
- Pagan LU, Gomes MJ, Okoshi MP. Endothelial function and physical exercise. Arq Bras Cardiol. 2018;111(4):540–541. doi:10.5935/abc.20180211
- Godo S, Shimokawa H. Endothelial Functions. Arterioscler Thromb Vasc Biol. 2017;37(9):e108–e114. doi:10.1161/ATVBAHA.117.309813
- Müller MM, Griesmacher A. Markers of endothelial dysfunction. Clin Chem Lab Med. 2000;38(2):77–85. doi:10.1515/CCLM.2000.013
- Meurice T, Vallet B, Bauters C, Dupuis B, Lablanche JM, Bertrand ME. Role of endothelial cells in restenosis after coronary angioplasty. Fundam Clin Pharmacol. 1996;10(3):234–242. doi:10.1111/j.1472-8206.1996.tb00302.x
- Darsaut T, Bouzeghrane F, Salazkin I, et al. The effects of stenting and endothelial denudation on aneurysm and branch occlusion in experimental aneurysm models. J Vasc Surg. 2007;45(6):1228–1235. doi:10.1016/j.jvs.2007.02.060
- Wang C, Wu H, Xing Y, et al. Endothelial-derived extracellular microRNA-92a promotes arterial stiffness by regulating phenotype changes of vascular smooth muscle cells. Sci Rep. 2022;12(1):344. doi:10.1038/s41598-021-04341-1
- Versari D, Lerman LO, Lerman A. The importance of reendothelialization after arterial injury. Curr Pharm Des. 2007;13(17):1811–1824. doi:10.2174/138161207780831239
- Bedair TM, ElNaggar MA, Joung YK, Han DK. Recent advances to accelerate re-endothelialization for vascular stents. J Tissue Eng. 2017;8:2041731417731546. doi:10.1177/2041731417731546
- Asahara T, Bauters C, Pastore C, et al. Local delivery of vascular endothelial growth factor accelerates reendothelialization and attenuates intimal hyperplasia in balloon-injured rat carotid artery. Circulation. 1995;91(11):2793–2801. doi:10.1161/01.CIR.91.11.2793
- Chen Y, Cao J, Peng W, Chen W. Neurotrophin-3 accelerates reendothelialization through inducing EPC mobilization and homing. Open Life Sci. 2020;15(1):241–250. doi:10.1515/biol-2020-0028
- Lüsebrink E, Goody PR, Lahrmann C, et al. AIM2 stimulation impairs reendothelialization and promotes the development of atherosclerosis in mice. Front Cardiovasc Med. 2020;7:582482. doi:10.3389/fcvm.2020.582482
- He J, Liu X, Su C, et al. Inhibition of mitochondrial oxidative damage improves reendothelialization capacity of endothelial progenitor cells via SIRT3 (Sirtuin 3)-enhanced SOD2 (superoxide dismutase 2) deacetylation in hypertension. Arterioscler Thromb Vasc Biol. 2019;39(8):1682–1698. doi:10.1161/ATVBAHA.119.312613
- Breen DM, Chan KK, Dhaliwall JK, et al. Insulin increases reendothelialization and inhibits cell migration and neointimal growth after arterial injury. Arterioscler Thromb Vasc Biol. 2009;29(7):1060–1066. doi:10.1161/ATVBAHA.109.185447
- Wara AK, Manica A, Marchini JF, et al. Bone marrow-derived Kruppel-like factor 10 controls reendothelialization in response to arterial injury. Arterioscler Thromb Vasc Biol. 2013;33(7):1552–1560. doi:10.1161/ATVBAHA.112.300655
- Urao N, Okigaki M, Yamada H, et al. Erythropoietin-mobilized endothelial progenitors enhance reendothelialization via Akt-endothelial nitric oxide synthase activation and prevent neointimal hyperplasia. Circ Res. 2006;98(11):1405–1413. doi:10.1161/01.RES.0000224117.59417.f3
- Li J, Chen Y, Gao J, et al. Eva1a ameliorates atherosclerosis by promoting re-endothelialization of injured arteries via Rac1/Cdc42/Arpc1b. Cardiovasc Res. 2021;117(2):450–461. doi:10.1093/cvr/cvaa011
- Xu H, Wu Z, Jin Z, et al. Paeonol suppresses vasculogenesis through regulating vascular smooth muscle phenotypic switching. J Endovasc Ther. 2022;29(1):117–131. doi:10.1177/15266028211032956
- Marin ML, Gordon RE, Veith FJ, Tulchin N, Panetta TF. Distribution of c-myc oncoprotein in healthy and atherosclerotic human carotid arteries. J Vasc Surg. 1993;18(2):170–176; discussion 176–177. doi:10.1016/0741-5214(93)90596-E
- Baudino TA, McKay C, Pendeville-Samain H, et al. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 2002;16(19):2530–2543. doi:10.1101/gad.1024602
- Bennett MR, Anglin S, McEwan JR, Jagoe R, Newby AC, Evan GI. Inhibition of vascular smooth muscle cell proliferation in vitro and in vivo by c-myc antisense oligodeoxynucleotides. J Clin Invest. 1994;93(2):820–828. doi:10.1172/JCI117036
- Florea V, Bhagavatula N, Simovic G, Macedo FY, Fock RA, Rodrigues CO. c-Myc is essential to prevent endothelial pro-inflammatory senescent phenotype. PLoS One. 2013;8(9):e73146. doi:10.1371/journal.pone.0073146
- Wang Y, Xu Y, Yan S, et al. Adenosine kinase is critical for neointima formation after vascular injury by inducing aberrant DNA hypermethylation. Cardiovasc Res. 2021;117(2):561–575. doi:10.1093/cvr/cvaa040
- Wang Y, Hu G, Liu F, et al. Deletion of yes-associated protein (YAP) specifically in cardiac and vascular smooth muscle cells reveals a crucial role for YAP in mouse cardiovascular development. Circ Res. 2014;114(6):957–965. doi:10.1161/CIRCRESAHA.114.303411
- Hu W, Wu X, Jin Z, et al. Andrographolide promotes interaction between endothelin-dependent EDNRA/EDNRB and myocardin-SRF to regulate pathological vascular remodeling. Front Cardiovasc Med. 2021;8:783872. doi:10.3389/fcvm.2021.783872
- Pang K, Dong Y, Hao L, et al. ERH interacts with EIF2alpha and regulates the EIF2alpha/ATF4/CHOP pathway in bladder cancer cells. Front Oncol. 2022;12:871687. doi:10.3389/fonc.2022.871687
- Gomez-Salinero JM, Rafii S. Endothelial cell adaptation in regeneration. Science. 2018;362(6419):1116–1117. doi:10.1126/science.aar4800
- Dejana E, Hirschi KK, Simons M. The molecular basis of endothelial cell plasticity. Nat Commun. 2017;8:14361. doi:10.1038/ncomms14361