Abstract
Proteomic approaches are continuing to make headways in cancer research by helping to elucidate complex signaling networks that underlie tumorigenesis and disease progression. This review describes recent advances made in the proteomic discovery of drug targets for therapeutic development. A variety of technical and methodological advances are overviewed with a critical assessment of challenges and potentials. A number of potential drug targets, such as baculoviral inhibitor of apoptosis protein repeat-containing protein 6, macrophage inhibitory cytokine 1, phosphoglycerate mutase 1, prohibitin 1, fascin, and pyruvate kinase isozyme 2 were identified in the proteomic analysis of drug-resistant cancer cells, drug action, and differential disease state tissues. Future directions for proteomics-based target identification and validation to be more translation efficient are also discussed.
Introduction
The use of proteomics as a systems biology tool in cancer research continues to expand in scope and depth, as it evolves rapidly into a universally applicable method for the investigation of practically any biological process. Proteomics is particularly attractive to cancer research because the complexity of tumorigenesis, cancer progression, tumor relapse, and metastasis often involves large protein networks. Indeed, in a little over a decade, applications of mass spectrometry-based proteomics in cancer-related research – ranging from mechanistic investigation to the discovery of novel therapeutic targets – have increased exponentially. This is evidenced by the explosive growth in the number of publications on the subject matter, from 109 in 2000 to 1,349 in 2011 (http://www.ncbi.nlm.nih.gov/pubmed), with the total number of publications containing the keywords “proteomics” and “cancer” reaching over 7,500 to date. A large number of review articles have appeared in the past several years, offering excellent overviews and perspectives on novel proteomic applications in cancer. Many reviews focused on different cancer types, such as breast cancer,Citation1–Citation4 pancreatic cancer,Citation5,Citation6 ovarian cancer,Citation7–Citation9 colorectal cancer,Citation10,Citation11 and glioma.Citation12–Citation14 Others have focused on sample types or subcellular components, such as tissue,Citation15–Citation17 serum,Citation18–Citation20 and secretome.Citation21–Citation23 In addition, other reviews focused on the innovative proteomic methodologies or the various end goals of proteomics in cancer, such as phosphoproteomics,Citation24–Citation27 mechanisms of drug resistance,Citation28–Citation30 and kinome profiling.Citation31–Citation33 Finally, there appear to be more reviews on proteomic biomarker discovery for cancer diagnosis, prognosis, and therapeutic response than on any other area of focus, highlighting the intense activities in the search for new biomarkers that could be translated to clinical applications.Citation30,Citation34–Citation36
Despite the remarkable advances in utilizing proteomics in almost every aspect of cancer research, many challenges remain. Global proteomic approaches, while capable of identifying thousands of proteins (many of which are differentially expressed) are often biased towards soluble and high-abundance proteins, thus easily missing out on many low-abundance proteins that could play critical roles in signal transduction.Citation37 Most solid tumors are heterogeneous, making it difficult to identify key proteins that could be used as universal targets even in the same type of cancer.Citation38,Citation39 While high-throughput proteomic methods can discover “unbiased” cohorts of potential biomarkers and therapeutic targets that regulate disease progression, the sheer number (often in the dozens, if not the hundreds) of hits could be cumbersome, as current functional studies are limited to involving one or a few proteins at a time. Thus, a large number of diagnostic and prognostic biomarkers have been identified by proteomic analysis, but very few have made it to the clinical stage, underscoring the urgent need for the convergence of functional studies and clinical sample validations. Moreover, the progress in drug development has far lagged the pace of proteomic discovery of potential drug targets, as the former generally requires a prolonged developmental process from drug design to in vitro and in vivo tests for efficacy and toxicities, as well as for absorption, distribution, metabolism, and excretion (ADME).Citation40
Proteomic approaches have been increasingly used in the preclinical drug discovery process to understand the effects of drug candidates on their protein targets and to shed light on the cellular mechanisms behind the observed phenotype, although many other biomaterials including deoxyribonucleic acid (DNA), ribonucleic acid (RNA), and carbohydrates can also be used as novel drug targets with different approaches.Citation41 Proteomic studies with the main goal of identifying new druggable targets for the treatment of cancer have been relatively few and are the primary focus of this review. For the convenience of discussion, we will attempt to group various proteomic target discovery studies into several distinct approaches for target identification.
Current proteomic techniques for target discovery
While mass spectrometry (MS)-based proteomic approaches remain the most comprehensive and versatile tool in large-scale proteomic profiling,Citation42,Citation43 several non-MS-based techniques, such as reverse phase protein microarrays (RPMA)Citation44 and peptide arraysCitation45,Citation46 have recently gained popularity. RPMA is a technology platform designed for the quantitative analysis of specific phosphorylated, cleaved, or total (phosphorylated and nonphosphorylated) forms of cellular proteins from complex mixtures in multiple samples simultaneously. One microarray can accommodate a range of hundreds to thousands of samples that are printed in a series of replication. RPMA is performed using either a primary or a secondary labeled antibody by chemiluminescent, fluorescent, or colorimetric assays. Multiplexing is also achieved by simultaneously probing multiple arrays spotted with the same lysate with different antibodies and can be implemented as a quantitative calibrated assay.Citation47 RPMA has now been utilized for potential drug discovery/validation, as well as for advancements in understanding the disease both in the clinic and in the laboratory,Citation48,Citation49 and RPMA is currently being integrated into human clinical cancer trials.Citation50
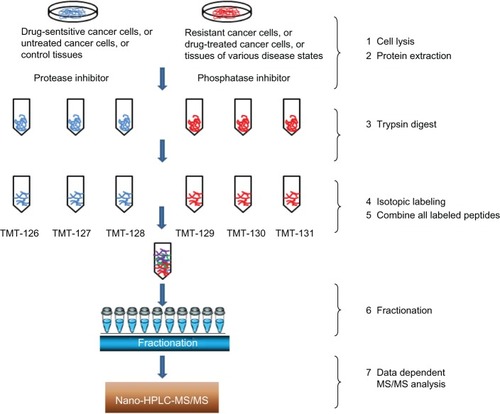
The quantitative methods adopted in MS include techniques based on the stable isotope labeling of proteins/peptides, as well as label-free methods. In comparison with labeling methods, label-free methods provide higher dynamic ranges of quantification and are versatile tools that are used to estimate changes in protein abundances between different samples.Citation51–Citation53 The utility of MS-based proteomic analyses and their applications in drug target identification have been increasingly recognized over the past decade due to their high sensitivity, specificity, and throughput. For example, gel-free isotopic labeling methods, such as SILAC (stable isotope labeling with amino acids in cell culture),Citation54,Citation55 isobaric tags for relative and absolute quantitation (iTRAQ),Citation56–Citation58 and isobaric tandem mass tags (TMTs)Citation59 () can now routinely quantify several hundreds of (to a few thousand) proteins in a single analysis with high reproducibility. Continued innovations and improvements in the instrumentation and bioinformatics tools will drive MS-based proteomics applications in drug discovery,Citation60,Citation61 complemented by streamlined and focused non-MS-based methods for targeted and repeated assays, such as protein arrays. The former will play key roles in the discovery stage, where quantitative proteomic analysis is undertaken, which compares differentially expressed proteins for potential drug targets. The array methods will be more cost effective and convenient to use for the validation of studies on drug targets and their effects.
Target identification in resistant cancer population
One of the greatest challenges in cancer therapy is drug resistance that occurs either intrinsically or is acquired after a certain period of treatment. Resistance to targeted cancer drugs involves complex and diverse molecular adaptations by cancer cells under the selective pressure of therapeutic regimens. For example, the specific target protein in tumors may undergo mutations to become inaccessible to drugs.Citation62–Citation64 In addition, a subset of cancer stem cells may be resistant to the cytotoxicity of drugs that target the bulk of more differentiated cancer cells.Citation65–Citation68 Moreover, chemotherapeutic agents often induce adaptive changes in the regulatory networks and signaling pathways, leading to the independence of cancer cells on the targeted protein and to the emergence of drug-resistant disease clones.Citation69 Indeed, such diverse mechanisms of drug resistance can transform cancer cells under the selective pressure of therapies, which made it extremely difficult to combat tumor recurrence; this is inevitably followed by metastatic progression of the disease and death. Current cancer treatment typically consists of a monotherapy regimen targeting a specific protein or signaling pathway, (for example, selective estrogen receptor modulators [SERMs] for the estrogen receptor, Herceptin for Her2, and gefitinib for epidermal growth factor receptor [EGFR]). However, tumors that are initially responsive eventually develop resistance to therapy via a variety of mechanisms. Switching to alternative drugs after the emergence of resistant disease clones following first-line treatment often achieves little in delaying cancer progression. Resistance can develop to just one drug, or tumors may be cross-resistant to several structurally dissimilar and functionally distinct agents, a phenomenon known as multidrug resistance.Citation70 Thus, there is an urgent need to better understand the resistance mechanism and to identify novel targets for a therapeutic intervention that can be more effective in blocking the progression of cancer cells already resistant to the previous treatment.
Here, proteomic approaches hold the promise to identify individual proteins and interactive signaling networks that act individually or collectively in conferring drug resistance. The comparison of protein expression between drug-resistant tumor tissues or cell lines with drug-sensitive tumor tissues or cell lines is one of the most promising tools for drug target identification. Targeting these proteins and signaling pathways may either resensitize tumor response to the original treatment, or it may help overcoming acquired drug resistance by blocking alternative survival signaling induced by the initial treatment. More importantly, proteomic characterization of a resistant phenotype can provide a more complete picture of signaling adaptation, as multiple drug targets for combinatorial therapeutic solutions can be identified to combat resistance and achieve longer-term improvement in disease outcomes. While advances in this application have been limited, the following examples demonstrate that proteomic approaches are inherently suitable for the discovery of targets for resistant phenotypes, and we expect an increasing number of protein targets to be validated by functional studies followed by therapeutic development.
One example comes from a comparative proteomic analysis of colon cancer stem cells and differentiated tumor cells, which uncovered a prominently upregulated protein, baculoviral inhibitor of apoptosis protein (IAP) repeat-containing protein 6 (BIRC6), one of the IAPs that may play a crucial role in the chemoresistance of colon cancer stem cells. Subsequent knockdown of the gene resulted in resensitization of the cells to chemotherapy.Citation71 Results from this proteomic study suggest that BIRC6 could be used as a potential therapeutic target to eradicate colon cancer stem cells contributing to colon cancer recurrence. In other cases, an iTRAQ–MS-based proteomics analysis was used to identify proteins associated with the development of docetaxel resistance by comparing docetaxel-sensitive PC3 cells and docetaxel-resistant PC3-Rx cells developed by docetaxel dose escalation.Citation56 Functional validation experiments were performed using recombinant protein treatment and small interfering (si)RNA knockdown experiments. The findings from this study suggested that macrophage inhibitory cytokine 1 should be further investigated as a potential biomarker and therapeutic target for acquired resistance to docetaxel treatment. However, extensive functional studies and follow-up validations are required before these proteins can be proven as viable drug targets.
Similarly, a quantitative proteomic investigation of hepatocellular carcinoma, a type of liver cancer known for its resistance to chemotherapy, implicated phospho-glycerate mutase 1 (PGAM1) as a potential therapeutic target.Citation72 Clinicopathological analysis indicated that the overexpression of PGAM1 was associated with 66.7% of hepatocellular carcinomas, and is strongly correlated with poor differentiation and decreased survival rates (P<0.01). In addition, short hairpin (sh)RNA-mediated repression of PGAM1 expression resulted in significant inhibition in liver cancer cell growth both in vitro and in vivo. Using a proteomics approach, elevated levels of prohibitin 1 (PHB1) and glutathione S-transferase pi (GSTπ) were found to be associated with paclitaxel resistance in discrete subcellular fractions of two drug-resistant sublines relative to their sensitive sublines.Citation73 Immunofluorescent staining and fractionation studies revealed an increased level of PHB1 on the surface of resistant cell lines. Transiently silencing either PHB1 or GSTπ gene expression using siRNA in the paclitaxel-resistant cancer cell sublines partially sensitized these cells toward paclitaxel. Intriguingly, silencing PHB1, but not GSTπ, resulted in the activation of intrinsic apoptosis pathway in response to paclitaxel. Similarly, stably silencing either PHB1 or GSTπ significantly improved paclitaxel sensitivity in A549TR cells both in vitro and in vivo. This study suggests that PHB1 is a mediator of paclitaxel resistance, and this resistance may depend on the cellular localization of proteins. Based on these preliminary functional studies, PHB1 is proposed as a potential target for therapeutic strategies for the treatment of drug-resistant tumors. Again, we have yet to see follow-up investigations of the feasibility of such novel, yet unproven, therapeutic targets.
In our recent proteomic study of letrozole-resistant breast cancer, a TMT label was used for a quantitative comparison of protein expression profiles of the resistant versus sensitive MCF-7 cells overexpressing aromatase.Citation74 This study identified fascin, among other significantly upregulated proteins, as a promising therapeutic target for the inhibition of metastasis of hormone-resistant breast cancer cells that have acquired the enhanced capacity of migration and invasion. Retrospective clinical validation confirms fascin overexpression as an independent indicator of decreased survival and poor prognosis. A subsequent drug discovery effort by our group led to the discovery of a series of thiazole compoundsCitation75 as potent inhibitors of migration and invasion of metastatic cancer cells by binding to fascin.
In summary, a resistant cancer population, whether inherent or acquired, is a major problem that reduces the activity of conventional and/or molecularly-targeted cancer drugs. The success of identifying drug targets in resistant cancer cells by proteomic approaches depends on reliable drug-resistant tumor tissues or cell lines. It is also critical that proteomics be combined with data from other approaches in an attempt to investigate mechanistic pathways in more detail, as well as to validate potential targets in a clinical setting.
Target identification from drug action (chemical proteomics)
Chemical proteomics is a technique that identifies proteins enriched or isolated as a result of interacting with or binding to a chemical probe, usually a small molecule drug that is fixed on a solid support.Citation76–Citation78 Therefore, these affinity-based enrichment techniques, in combination with MS, have enabled the direct determination of protein-binding profiles of small molecule drugs under physiological conditions and represent one of the most direct approaches to screen for drug–protein interactions.Citation79,Citation80 The major drawback encountered in the affinity-based chemical proteomics is the presence in the pulled-down extract of nonspecifically bound proteins. Among these, a large number of proteins that bind nonspecifically to most conventional affinity matrices have been reported.Citation81 It is, therefore, strongly recommended that green fluorescent protein be used as the tag of choice because it shows minimal nonspecific binding to mammalian cell proteins, it can be quantitatively depleted from cell extracts, and it allows for the integration of biochemical protein interaction data with in vivo measurements using fluorescence microscopy. In addition, it is also necessary to perform appropriate negative control experiments to distinguish nonspecific interactions from specific interactions. Chemical proteomic approaches also include global proteomic profiling of cellular samples treated with a biologically active compound without enrichment steps. In pharmacology, chemical proteomics have been utilized to determine the specificity of drugs and their analogs, for anticipated as well as unknown targets that may also bind to the probe.Citation41 These activity-based probes can specifically target diverse sets of enzyme families and provides direct information about the activation state of identified proteins.Citation82 Several kinds of chemical probes have been used in proteomic studies across a multitude of enzyme classes such as hydrolases, proteases, kinases, phosphatases, histone deacetylases, glycosidases, and oxidoreductases.Citation83 Thus, chemical proteomic approaches not only identify protein targets for drugs that exert known biological activities in vitro or in vivo, but they also can discover previously unknown targets for drugs of known modes of action (MoA). These off-target proteins may be additional druggable targets, but they may also account for the side effects and toxicities. Identification of protein targets that are responsible for toxicities may prove valuable in early drug development to minimize failure in clinical trials. In addition, chemical proteomics can identify novel drug targets by broad-action chemical probes (for example, nonselective kinase inhibitors such as Staurosporine [Life Technologies, Carlsbad, CA, USA]).Citation82 In the following, we present a few examples in which chemical proteomics were used effectively for target discovery and validation.
Tyrosine kinase inhibitors represent a large group of targeted therapeutics for cancer. Gefitinib (Iressa™; AstraZenca, London, UK) is the first selective inhibitor of the EGFR tyrosine kinase domain.Citation84 Gefitinib used as monotherapy is an effective treatment for patients with locally advanced or metastatic nonsmall cell lung cancer (NSCLC) with EGFR mutations.Citation85 Gefitinib has also been tested in clinical trials in other tumors, including head and neck squamous cell carcinoma (HNSCC), as monotherapy, or in combination with other chemotherapies or radiation, but it has shown limited clinical efficacy with response rates of 10%–15%.Citation86–Citation89 Using cleavable isotope-coded affinity tagging (cICAT)-based liquid chromatography–tandem mass spectrometry (LC–MS/MS) method, Chen et alCitation90 identified the tyrosine phosphorylation levels of 21 proteins between control and epidermal growth factor-treated A431 human cervical cancer cells. Of these, endofin, DCBLD2, and KIAA0582 were validated to be novel tyrosine-phosphorylation targets of epidermal growth factor signaling and responsive to gefitinib therapy.Citation90 Pernas et alCitation91 observed that gefitinib sensitivity correlated with phospho (p)-AKT and p-STAT3 activation in HNSCC cell lines and tumor specimens, thus p-AKT and p-STAT3 could serve as potentially useful drug targets for further development of novel therapeutic agents for HNSCC. More recently, Wu et alCitation92 used quantitative chemical proteomics to identify several kinases including NEK9, LYN, JAK1, WEE1, and EPHA2, which are involved in cell survival and the proliferation of HNSCC cell lines. These findings may lead to new therapeutic options for HNSCC patients.
For inhibitors acting on multiple tyrosine kinases, chemical proteomic studies could help uncover additional unknown targets and their interacting signaling network. For example, as an inhibitor of multiple tyrosine kinases, dasatinib targets Bcr-Abl, SRC family kinases, c-Kit, and platelet-derived growth factor receptor kinase, a cell-permeable kinase probe (DA-2) was employed to profile potential cellular targets of dasatinib; a number of previously unknown dasatinib targets were identified, including several serine/threonine kinases (PCTK3, STK25, eIF-2A, PIM-3, PKA C-α, and PKN2).Citation93 According to another report,Citation94 nearly 40 different kinase targets of dasatinib were discovered using quantitative phosphoproteomics. These include receptor tyrosine kinases (ephrin receptors, discoidin domain receptor 1, and EGFR) and nonreceptor tyrosine kinases (FRK, BRK, and ACK). These results provided a system-level view of dasatinib action in cancer cells and suggested functional targets and rationales for therapeutic strategies.
When the mechanisms of biologically active compounds against cancer cells are not fully understood, chemical proteomics could help elucidate the modes of action by mapping out protein networks perturbed by the drug treatment. In this aspect, a wealth of proteomic discoveries have seldom been translated into validated drug mechanisms, most of which stayed as preliminary findings that could lead to confirmed targets by in-depth functional studies. For example, celecoxib (Celebrex®; Pfizer, Inc, New York, NY, USA), originally a widely prescribed nonsteroidal anti-inflammatory drug, has recently been shown to have anticancer propertiesCitation95,Citation96 with uncertain mechanisms, ranging from modulating the expression of Bcl-2 family members and mitochondria-mediated apoptosis,Citation95,Citation97 to inhibiting nuclear factor kappa B,Citation98,Citation99 Akt,Citation100 and Stat3101signaling pathways. Proteomic analysis of human oral squamous cell carcinoma found that celecoxib treatment induced ten- to 20-fold overexpression of heterogeneous nuclear ribonuclear protein C.Citation102 Similarly, global proteomic profiles of colorectal cancer cells before and after treatment with celecoxib revealed significant alterations among multiple proteins involved in diverse cellular functions ranging from glycolysis, protein biosynthesis, DNA synthesis, messenger RNA processing, protein folding, phosphorylation, redox regulation, to molecular chaperon activities.Citation103 However, none of the proteomic studies has been conclusive in the mechanistic interpretation of numerous alterations in protein expression.
The Wnt-β-catenin signaling pathway is a developmental signaling pathway that plays a critical role in regulation, differentiation, proliferation, and apoptosis; thus, aberrant Wnt-β-catenin signaling is widely implicated in numerous cancers.Citation104–Citation106 However, identification of targeted Wnt-β-catenin pathway inhibitors in cancer patient treatment has been hampered by the limited number of pathway components that are amenable to small molecule inhibition. Recently, Huang et alCitation107 used an iTRAQ approach to identify a small molecule, XAV939, that selectively inhibits β-catenin-mediated transcription. Interestingly, XAV939 also inhibits the poly-adenosine diphosphate-ribosylating enzymes, tankyrase 1 and tankyrase 2, which interact with a highly conserved domain of axin and stimulate its degradation through the ubiquitin–proteasome pathway.Citation107 Tankyrases, involved in fundamental cellular processes such as telomere homeostasis and Wnt signaling, are potential telomere-directed anticancer targets.Citation108,Citation109 Therefore, XAV939 is not only a useful tool in understanding Wnt-β-catenin signaling, but also a potential drug that targets Wnt-β-catenin and telomeres.
Chemical proteomics aimed at characterizing the effects of drug candidates could sometimes lead to the discovery of new drug targets. For example, sulforaphane (SFN) is known to have antimicrobial and anticancer properties in experimental models.Citation110–Citation112 SFN can modulate multiple cellular targets involved in cancer development including DNA protection, inhibition of cancer cell proliferation, induction of apoptosis, inhibition of neoangiogenesis, progression of benign tumors to malignant tumors, and metastasis.Citation112,Citation113 Mastrangelo et alCitation114 identified serotonin receptors as the novel targets of SFN by proteomic analysis in Caco-2 colon cancer cells. This finding may not only shed light on the serotonin-mediated signaling pathways in colon cancer, but it may also lead to the development of potential novel therapeutic agents targeting serotonin. In another study, phosphoglucomutase 3 was identified by proteomic analysis and may contribute to SFN-induced cell death in the LNCaP prostate cancer (PCa) cells, which make phosphoglucomutase 3 a potential molecular therapeutic target for PCa.Citation115 While these are very early studies on the discovery of druggable protein targets in the relevant disease, they do open up new venues for the development of novel treatment regimens.
Although the field of chemical proteomics has proven its value in identifying novel drug targets, several challenges still remain to be overcome. First, the chemical probes must be carefully designed to covalently attach to proteins of interest and allow purification and/or identification. Second, the spectrum of available activity-based probes needs to be broadened in order to target additional enzyme classes. In addition, the development of high-throughput and gel-free assays, in conjunction with activity-based probes, will be required to enhance the experimental value of chemical proteomics. The continued success of chemical proteomics depends on the design of novel probes or new probe classes that can specifically target diverse sets of enzyme families, as well as on an unbiased assessment of the full spectrum of drug–target interactions and their molecular MoA.
Target discovery based upon differential disease state tissues (tissue proteomics)
Proteins and their spatiotemporal distribution changes play a central role in biological processes, including cancer initiation and progression. Analysis of a specific homogeneous cell type from tumor tissues could reveal molecular changes that take place in tumorigenesis since the concentration of disease-related proteins are likely to be much higher within or near the tumor areas. Recently, large-scale protein identification and comparative quantitation of highly complex protein mixtures have been achieved with proteomic strategies based upon differential disease state tissues, known as tissue proteomics.Citation35,Citation116–Citation120 Remarkable advances in tissue proteomics have been propelled by the rapid development of efficient methodologies and techniques in innovative sample preparation, sophisticated MS instrumentation, and powerful bioinformatics tools. The goals of tissue proteomic research focus on early or more accurate diagnosis, improvement of therapeutic strategies, and better evaluation of prognosis and/or prevention of a given disease, as well as on the identification of novel drug targets based upon the differential protein expression between “control” and “case” groups for the disease. A major hurdle in tissue proteomics analysis is the variability observed among the tissue samples due to the heterogeneity of tumor tissues containing cancer cells, as well as inflammatory, vascular, and connective tissue cells. Detailed proteomic analyses of these clinically valuable samples require meticulous preparation procedures, which is a major current focus of tissue proteomics. So far, tissue proteomics has uncovered large numbers of proteins with altered expressions in tumor tissues by analyzing both fresh–frozen biopsy samples and archived tumor tissues stabilized with formalin fixation and paraffin embedding (FFPE). However, very few, if any, of these proteins have actually become novel diagnostic biomarkers or therapeutic targets. As illustrated in the examples below, most tissue proteomic studies remain as preliminary investigations that identified differentially expressed proteins in diseased tissues. While these proteins could be potential prognostic markers or therapeutic targets, there is a general lack of in-depth functional and validated studies that follow up on these initial screening results.
Tissue proteomics has been employed quite frequently in the study of lung cancer for molecular mechanism elucidation as well as for novel drug target discovery.Citation121–Citation126 Peng et alCitation123 studied the protein profile changes between human pulmonary adenocarcinoma tissues and paired surrounding normal tissue with two-dimensional (2D) gel electrophoresis and ESI-Q-TOF (electrospray ionization, quadrupole, time-of-flight) MS/MS instruments. Thirty-two differentially expressed proteins (>2-fold change; P<0.05) were identified in pulmonary adenocarcinoma when compared to normal tissues. Knockdown of pyruvate kinase isozyme 2 (PKM2), one of the overexpressed proteins, led to a significant suppression of cell growth, to the induction of apoptosis in vitro, and to tumor growth inhibition in vivo. Moreover, the shRNA-expressing plasmid targeting cofilin-1, another over-expressed protein, significantly inhibited tumor metastases and prolonged survival in vivo. This tissue proteomic analysis and validation indicated that PKM2 and cofilin-1 could be potential therapeutic targets for pulmonary adenocarcinoma. It should be noted that small molecule inhibitors of PKM2 were identified and shown to inhibit cancer cell glycolysis and increase cancer cell death following the loss of growth factor signaling,Citation127 which was consistent with the proteomics-driven conclusion. Cofilin-1 has long been associated with increased tumor metastasis due to its role in regulating cytoskeleton dynamics,Citation128 and the inhibition of cofilin-1 by small molecule inhibitors has been demonstrated to enhance actin depolymerization.Citation129 In another example, Carretero et alCitation124 performed an integrated genomic and proteomic study for the identification of genes and phosphoprotein status associated with Lkb1 loss and progression to invasive and metastatic lung tumors in primary and metastatic de novo lung cancers. Phosphoproteomic analysis determined that two key modulators of focal adhesion dynamics, Src and focal adhesion kinase, are upregulated by Lkb1 loss during NSCLC progression. Moreover, the combined inhibition of Src, phosphoinositide 3-kinase, and MEK1/2 resulted in a synergistic tumor regression. These results point towards a mechanism underlying the increased propensity for metastases seen in Lkb1-deficient lung tumors, and they identified the Src signaling pathway as a molecularly targetable pathway for the treatment of LKB1-deficient NSCLC in humans. Therefore, development of therapeutic Src inhibitors, including dasatinib and saracatinib, will be a valid therapeutic strategy for the treatment of LKB1-deficient NSCLC.
Proteomic analysis of PCa tissues associated with multistage tumor progression also provides a valuable source of clinically relevant biomarkers and novel therapeutic targets.Citation130,Citation131 For example, Ummanni et alCitation132 recently reported the differential protein expression patterns from histologically characterized PCa tumor tissues and surrounding benign tissues of individual PCa patients based upon 2D differential gel electrophoresis coupled with MS. The study identified 118 protein spots differentially expressed in cancer (n=24) when compared to benign (n=21) prostate tissues adjacent to cancerous tissues. Analysis of these gel spots by matrix-assisted laser desorption/ionization–time-of-flight MS/MS revealed 79 unique proteins. Moreover, system biology analysis of proteomic results revealed several novel drug targets of PCa development and/or progression including eIF4A3, DDAH1, ARG2, Prdx3, and Prdx4, although functional validation of individual targets have yet to be performed. It is evident that proteomic analysis of multistage PCa tissues could provide new insights into PCa progression and potentially lead to the design of novel diagnostic and therapeutic strategies.
Efficient peptide/protein extraction approaches are crucial to the success of tissue proteomic analysis. Two major strategies of mining proteomic information from FFPE archive tissue samples have been developed.Citation133–Citation138 One strategy aimed to recover full-length proteins by heat treatment in suitable buffers with consequent reversion of the formaldehyde-induced cross-links. The operation conditions have been optimized by various labs since Shi et alCitation139 initially established this method. Application of high temperatures and the addition of detergent sodium dodecyl sulfate were indicated as two critical conditions for enhanced protein extraction yields.Citation140–Citation142 Because the protein was deposited in large amounts as insoluble, densely packed aggregates, applications of 40,000 psi pressure were reported to recover 96% of proteins from a tissue surrogate model, compared with a 26% recovery rate at 14.7 psi.Citation138,Citation143 Another major strategy is based upon the direct proteolytic digestion of intact FFPE archive tissue samples, followed by liquid chromatography–MS/MS characterization of the complex peptide mixture.Citation144 Tissue solubilization can be achieved in various buffers including sodium dodecyl sulfate–dithiothreitol,Citation145 radioimmunoprecipitation assay,Citation146 acetonitrile ammonium bicarbonate,Citation147 Tris-HClCitation148 under different temperatures, and pH conditions for the optimal yield of peptides. As a successful example, Hwang et alCitation147 characterized 428 prostate-expressed proteins from FFPE archive tissue samples for the discovery of PCa biomarkers and potential drug targets using the shotgun approach. Results to date demonstrate that direct trypsin protein digestion is an effective sample preparation strategy for proteomic analysis of FFPE archive tissues.
Approximately 70% of diagnosed breast cancers express the estrogen receptor (ER+) whereas ER– breast cancers are not well differentiated and clinically tend to be more aggressive.Citation149–Citation152 A global proteomic characterization and quantitative comparison of ER+ and ER– breast tumors were recently performed on fresh–frozen breast tumor tissues.Citation153 The study identified 2,995 unique proteins including a number of receptor tyrosine kinases and intracellular kinases that are abundantly expressed in both ER+ and ER– breast cancer tissues. Using a label-free quantitative approach, 236 proteins were found differentially expressed between ER+ and ER– breast tumors. Compared with ER– breast tumors, 141 proteins were selectively upregulated, while 95 proteins were downregulated in ER+ tumors. Molecular function analysis of gene ontology showed that the dehydrogenase, reductase, cytoskeletal proteins, extracellular matrix, hydrolase, and lyase categories were significantly enriched in ER+ breast tumors, whereas selected calcium-binding proteins, membrane traffic proteins, and cytoskeletal proteins were enriched in ER– tumors. Biological process and pathway analysis indicated that proteins related to amino acid metabolism, proteasome, and fatty acid metabolism were overexpressed in ER+ tumors, while proteins related to the glycolysis pathway were overexpressed in ER– tumors. Given the clinical challenges in treating ER– breast cancer, such in vivo findings of differentially expressed proteins in ER– tumors are especially significant because of their pathological relevance. These proteins may serve as potential therapeutic targets for ER– breast cancer if functionally validated. Recently, Cabezón et alCitation154 used a systematic 2D gel-based proteomic profiling strategy, applied to the analysis of 78 fresh triple-negative breast cancer (TNBC) tissue biopsies, in combination with a three-tier orthogonal technology (2D polyacrylamide gel electrophoresis/silver staining coupled with MS, 2D Western blotting, and immunohistochemistry) approach, they identified and validated one specific protein, Mage-A4, which was expressed in a significant fraction of TNBC and Her2-positive/ER– lesions. The existence of immunotherapeutic approaches specifically targeting this protein, or Mage-A protein family members, provides novel management options for TNBC and Her2-positive/ER– patients bearing Mage-A4 positive tumors.
Tissue proteomics for cancer drug target identification is still in its infancy. Although promising results have been reported, there has yet to be a breakthrough that is close to becoming clinically applicable. The challenge will be to perform robustly designed, large retrospective studies including independent validation sets, followed by prospective validation studies to demonstrate the clinical benefits for patients. The development of siRNA in vivo techniques will greatly speed up selection of useful proteins. Characterization of discriminator proteins will provide new molecularly targeted anticancer drugs. Faced with the complexity of cancer tissue proteomics and its great clinical potential, a global coordination of ongoing efforts appears to be crucial.
Conclusion
In this review we provided an overview of three major approaches of proteomics employed in discovering potential therapeutic targets for cancer. Target identification in resistant cancer populations is one of the most promising tools for drug target identification, and it can uncover multiple drug targets for combinatorial therapeutic solutions to combat resistance and achieve longer-term improvement in disease outcomes. Chemical proteomics approaches not only identify protein targets for drugs that exert known biological activities, but they can also discover previously unknown targets for drugs of known MoA. The tissue proteomics approach offers the advantage of direct clinical relevance and can complement mechanism-based approaches like chemical proteomics.
Proteomics continues to evolve rapidly both in instrumental advancement and innovative applications in biomedical research, particularly in the field of cancer. The complexity of cancer biology requires not only in-depth functional studies for mechanistic elucidation, but also “unbiased” system approaches for a global view of interactive signaling networks that reflect the cancer disease stage. Cancer proteomics has uncovered a phenomenal number of cancer-specific proteomic alterations, shedding light on previously unknown mechanisms in tumorigenesis, cancer progression, and metastasis. Perhaps more prominently, proteomics has provided an unmatched wealth of potential biomarkers that could be clinically used for the diagnosis and prediction of treatment responses. The utility of proteomics as a valuable tool for the discovery of new cancer treatment targets has been increasingly recognized in recent years. We note the remarkable progress in several directions when cancer proteomics plays an important role in drug discovery, as illustrated in . Novel proteomics methodologies, both MS-based and non-MS-based, continued to be optimized and perfected using high-resolution and high-speed MS, simplified and robust array technologies for tumor tissue, cell, subcellular compartment, and other relevant biological samples. Proteomics has been increasingly used to elucidate the mechanisms of drug resistance in cancer with the hope that novel protein targets could emerge for therapeutic intervention of the resistant disease. In other innovative frontiers, chemical proteomics studies examining drug-induced biological perturbations often reveal new clinically relevant biomarkers and drug targets; in dissecting mechanisms of cancer progression, proteomics also identified previously unknown or unconsidered druggable targets.
Figure 2 Quantitative proteomic analysis in target discovery, validation, and drug development.
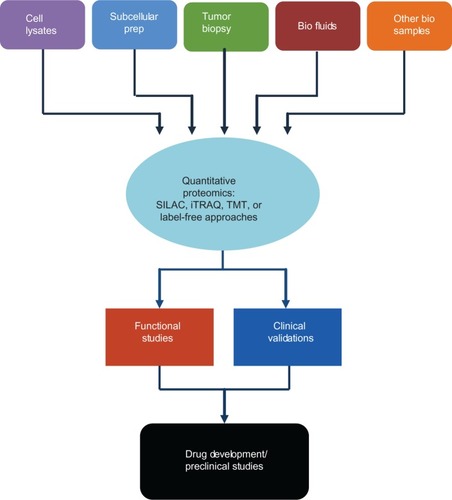
However, despite the tremendous progress and potential of proteomics for target discovery, there are serious challenges. A lack of coordinated efforts to follow up with the proteomic discovery of novel therapeutic targets largely accounts for the current gap between drug development (design, synthesis, and optimization) and new drug targets emerging from numerous proteomic studies of cancer. Most of the proteins identified in the proteomic analyses of drug resistance have not been validated for their roles in specific disease processes and their potential in clinical use. Functional studies involving in vitro manipulations of gene expression using specific pharmacological inhibitors, antisense RNA, RNA interference, or gene knockout experiments should be an integral part of a proteomic study for target discovery. Clinical validations of candidate biomarkers should also be included in retrospective and prospective studies. Future investigations should place more emphasis on functional studies and on the clinical validation of novel targets for their translation in clinical trials with greater speed and higher success rates. Proteomics is also expected to play a major role in the preclinical and clinical research of targeted and combinatorial therapies.
Acknowledgments
This work was funded in part by the National Institutes of Health/National Institute on Minority and Health Disparities grant number 8G12MD007590 (JZ) and 3G12MD007595 (GW), the Louisiana Cancer Research Consortium (GW), the Department of Agriculture Grant 58-6435-7-019 (GW), and the Office of Naval Research Grant N00014-99-1-0763 (GW).
Disclosure
The authors report no conflicts of interest in this work.
References
- Hondermarck H Tastet C El Yazidi-Belkoura I Toillon RA Le Bourhis X Proteomics of breast cancer: the quest for markers and therapeutic targets J Proteome Res 2008 7 4 1403 1411 18311906
- Fink-Retter A Czerwenka K Gschwantler-Kaulich D Proteomics in mammary cancer research Eur J Gynaecol Oncol 2009 30 6 635 639 20099493
- Griffith OL Gray JW Omic approaches to preventing or managing metastatic breast cancer Breast Cancer Res 2011 13 6 230 22216753
- Sharma N Martin A McCabe CJ Mining the proteome: the application of tandem mass spectrometry to endocrine cancer research Endocr Relat Cancer 2012 19 4 R149 R161 22555494
- Gräntzdörffer I Carl-McGrath S Ebert MP Röcken C Proteomics of pancreatic cancer Pancreas 2008 36 4 329 336 18437077
- Tonack S Aspinall-O’Dea M Neoptolemos JP Costello E Pancreatic cancer: proteomic approaches to a challenging disease Pancreatology 2009 9 5 567 576 19657212
- Faça VM Hanash SM In-depth proteomics to define the cell surface and secretome of ovarian cancer cells and processes of protein shedding Cancer Res 2009 69 3 728 730 19155298
- Hays JL Kim G Giuroiu I Kohn EC Proteomics and ovarian cancer: integrating proteomics information into clinical care J Proteomics 2010 73 10 1864 1872 20561909
- Callesen AK Mogensen O Jensen AK Reproducibility of mass spectrometry based protein profiles for diagnosis of ovarian cancer across clinical studies: A systematic review J Proteomics 2012 75 10 2758 2772 22366292
- Barderas R Babel I Casal JI Colorectal cancer proteomics, molecular characterization and biomarker discovery Proteomics Clin Appl 2010 4 2 159 178 21137041
- McHugh SM O’Donnell J Gillen P Genomic and oncoproteomic advances in detection and treatment of colorectal cancer World J Surg Oncol 2009 7 36 19338662
- Kalinina J Peng J Ritchie JC Van Meir EG Proteomics of gliomas: initial biomarker discovery and evolution of technology Neuro Oncol 2011 13 9 926 942 21852429
- Jung CS Unterberg AW Hartmann C Diagnostic markers for glioblastoma Histol Histopathol 2011 26 10 1327 1341 21870336
- Niclou SP Fack F Rajcevic U Glioma proteomics: status and perspectives J Proteomics 2010 73 10 1823 1838 20332038
- Cazares LH Troyer DA Wang B Drake RR Semmes OJ MALDI tissue imaging: from biomarker discovery to clinical applications Anal Bioanal Chem 2011 401 1 17 27 21541816
- Schwamborn K Gaisa NT Henkel C Tissue and serum proteomic profiling for diagnostic and prognostic bladder cancer biomarkers Expert Rev Proteomics 2010 7 6 897 906 21142890
- Moreira JM Cabezón T Gromova I Tissue proteomics of the human mammary gland: towards an abridged definition of the molecular phenotypes underlying epithelial normalcy Mol Oncol 2010 4 6 539 561 21036680
- Somasundaram K Nijaguna MB Kumar DM Serum proteomics of glioma: methods and applications Expert Rev Mol Diagn 2009 9 7 695 707 19817554
- Solassol J Du-Thanh A Maudelonde T Guillot B Serum proteomic profiling reveals potential biomarkers for cutaneous malignant melanoma Int J Biol Markers 2011 26 2 82 87 21607923
- Fanayan S Hincapie M Hancock WS Using lectins to harvest the plasma/serum glycoproteome Electrophoresis 2012 33 12 1746 1754 22740463
- Chen ST Pan TL Juan HF Chen TY Lin YS Huang CM Breast tumor microenvironment: proteomics highlights the treatments targeting secretome J Proteome Res 2008 7 4 1379 1387 18290608
- Xue H Lu B Lai M The cancer secretome: a reservoir of biomarkers J Transl Med 2008 6 52 18796163
- Stastna M Van Eyk JE Secreted proteins as a fundamental source for biomarker discovery Proteomics 2012 12 4–5 722 735 22247067
- Harsha HC Pandey A Phosphoproteomics in cancer Mol Oncol 2010 4 6 482 495 20937571
- Trost M Bridon G Desjardins M Thibault P Subcellular phosphoproteomics Mass Spectrom Rev 2010 29 6 962 990 20931658
- Ashman K Villar EL Phosphoproteomics and cancer research Clin Transl Oncol 2009 11 6 356 362 19531450
- Chong PK Lee H Kong JW Loh MC Wong CH Lim YP Phosphoproteomics, oncogenic signaling and cancer research Proteomics 2008 8 21 4370 4382 18814326
- Li XH Li C Xiao ZQ Proteomics for identifying mechanisms and biomarkers of drug resistance in cancer J Proteomics 2011 74 12 2642 2649 21964283
- Koomen JM Smalley KS Using quantitative proteomic analysis to understand genotype specific intrinsic drug resistance in melanoma Oncotarget 2011 2 4 329 335 21505227
- Hodgkinson VC Eagle GL Drew PJ Lind MJ Cawkwell L Biomarkers of chemotherapy resistance in breast cancer identified by proteomics: current status Cancer Lett 2010 294 1 13 24 20176436
- Piersma SR Labots M Verheul HM Jiménez CR Strategies for kinome profiling in cancer and potential clinical applications: chemical proteomics and array-based methods Anal Bioanal Chem 2010 397 8 3163 3171 20526883
- Parikh K Peppelenbosch MP Kinome profiling of clinical cancer specimens Cancer Res 2010 70 7 2575 2578 20332226
- Hochgräfe F Zhang L O’Toole SA Tyrosine phosphorylation profiling reveals the signaling network characteristics of Basal breast cancer cells Cancer Res 2010 70 22 9391 9401 20861192
- Liang S Xu Z Xu X Zhao X Huang C Wei Y Quantitative proteomics for cancer biomarker discovery Comb Chem High Throughput Screen 2012 15 3 221 231 22221055
- Lin LL Huang HC Juan HF Discovery of biomarkers for gastric cancer: a proteomics approach J Proteomics 2012 75 11 3081 3097 22498886
- Leth-Larsen R Lund RR Ditzel HJ Plasma membrane proteomics and its application in clinical cancer biomarker discovery Mol Cell Proteomics 2010 9 7 1369 1382 20382631
- Drake RR Cazares LH Jones EE Fuller TW Semmes OJ Laronga C Challenges to developing proteomic-based breast cancer diagnostics OMICS 2011 15 5 251 259 21332380
- Abramovitz M Leyland-Jones B Application of array-based genomic and epigenomic technologies to unraveling the heterogeneous nature of breast tumors: on the road to individualized treatment Cancer Genomics Proteomics 2007 4 3 135 145 17878517
- Lam SW Jimenez CR Boven E Breast cancer classification by proteomic technologies: Current state of knowledge Cancer Treat Rev Epub 7 23 2013 pii S0305-7372(13)00129-00121
- Ohtsuki S Uchida Y Kubo Y Terasaki T Quantitative targeted absolute proteomics-based ADME research as a new path to drug discovery and development: methodology, advantages, strategy, and prospects J Pharm Sci 2011 100 9 3547 3559 21560129
- Schirle M Bantscheff M Kuster B Mass spectrometry-based proteomics in preclinical drug discovery Chem Biol 2012 19 1 72 84 22284356
- Rajcevic U Niclou SP Jimenez CR Proteomics strategies for target identification and biomarker discovery in cancer Front Biosci (Landmark Ed) 2009 14 3292 3303 19273274
- Mikami T Aoki M Kimura T The application of mass spectrometry to proteomics and metabolomics in biomarker discovery and drug development Curr Mol Pharmacol 2012 5 2 301 316 22122469
- Creighton CJ Fu X Hennessy BT Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancer Breast Cancer Res 2010 12 3 R40 20569503
- Arsenault R Griebel P Napper S Peptide arrays for kinome analysis: new opportunities and remaining challenges Proteomics 2011 11 24 4595 4609 22002874
- Beyer M Block I König K A novel combinatorial approach to high-density peptide arrays Methods Mol Biol 2009 570 309 316 19649602
- Sheehan KM Calvert VS Kay EW Use of reverse phase protein microarrays and reference standard development for molecular network analysis of metastatic ovarian carcinoma Mol Cell Proteomics 2005 4 4 346 355 15671044
- Wilson B Liotta LA Petricoin E Monitoring proteins and protein networks using reverse phase protein arrays Dis Markers 2010 28 4 225 232 20534907
- Voshol H Ehrat M Traenkle J Bertrand E van Oostrum J Antibody-based proteomics: analysis of signaling networks using reverse protein arrays FEBS J 2009 276 23 6871 6879 19860827
- Sereni MI Pierobon M Angioli R Petricoin EF Frederick MJ Reverse phase protein microarrays and their utility in drug development Methods Mol Biol 2013 986 187 214 23436414
- Patel VJ Thalassinos K Slade SE A comparison of labeling and label-free mass spectrometry-based proteomics approaches J Proteome Res 2009 8 7 3752 3759 19435289
- Bantscheff M Schirle M Sweetman G Rick J Kuster B Quantitative mass spectrometry in proteomics: a critical review Anal Bioanal Chem 2007 389 4 1017 1031 17668192
- Megger DA Bracht T Meyer HE Sitek B Label-free quantification in clinical proteomics Biochim Biophys Acta 2013 1834 8 1581 1590 23567906
- Barbi de Moura M Vincent G Fayewicz SL Mitochondrial respiration – an important therapeutic target in melanoma PLoS One 2012 7 8 e40690 22912665
- Sharma K Vabulas RM Macek B Quantitative proteomics reveals that Hsp90 inhibition preferentially targets kinases and the DNA damage response Mol Cell Proteomics 2012 11 3 M111.014654
- Zhao L Lee BY Brown DA Identification of candidate biomarkers of therapeutic response to docetaxel by proteomic profiling Cancer Res 2009 69 19 7696 7703 19773444
- Yokoyama T Enomoto T Serada S Plasma membrane proteomics identifies bone marrow stromal antigen 2 as a potential therapeutic target in endometrial cancer Int J Cancer 2013 132 2 472 484 22729361
- Zhang J Nkhata K Shaik AA Mouse prostate proteome changes induced by oral pentagalloylglucose treatment suggest targets for cancer chemoprevention Curr Cancer Drug Targets 2011 11 7 787 798 21762084
- Dillon R Nilsson CL Shi SD Lee NV Krastins B Greig MJ Discovery of a novel B-Raf fusion protein related to c-Met drug resistance J Proteome Res 2011 10 11 5084 5094 21936566
- Altelaar AF Munoz J Heck AJ Next-generation proteomics: towards an integrative view of proteome dynamics Nat Rev Genet 2013 14 1 35 48 23207911
- Nesvizhskii Al Vitek O Aebersold R Analysis and validation of proteomic data generated by tandem mass spectrometry Nat Methods 2007 4 10 787 797 17901868
- Phillips KA Van Bebber S Issa AM Diagnostics and biomarker development: priming the pipeline Nat Rev Drug Discov 2006 5 6 463 469 16718275
- Hoskins JM Carey LA McLeod HL CYP2D6 and tamoxifen: DNA matters in breast cancer Nat Rev Cancer 2009 9 8 576 586 19629072
- Wheeler DL Dunn EF Harari PM Understanding resistance to EGFR inhibitors-impact on future treatment strategies Nat Rev Clin Oncol 2010 7 9 493 507 20551942
- Al-Ejeh F Smart CE Morrison BJ Breast cancer stem cells: treatment resistance and therapeutic opportunities Carcinogenesis 2011 32 5 650 658 21310941
- Gangemi R Paleari L Orengo AM Cancer stem cells: a new paradigm for understanding tumor growth and progression and drug resistance Curr Med Chem 2009 16 14 1688 1703 19442140
- Milas L Hittelman WN Cancer stem cells and tumor response to therapy: current problems and future prospects Semin Radiat Oncol 2009 19 2 96 105 19249647
- Frame FM Maitland NJ Cancer stem cells, models of study and implications of therapy resistance mechanisms Adv Exp Med Biol 2011 720 105 118 21901622
- Fodale V Pierobon M Liotta L Petricoin E Mechanism of cell adaptation: when and how do cancer cells develop chemoresistance? Cancer J 2011 17 2 89 95 21427552
- Thomas H Coley HM Overcoming multidrug resistance in cancer: an update on the clinical strategy of inhibiting p-glycoprotein Cancer Control 2003 10 2 159 165 12712010
- Van Houdt WJ Emmink BL Pham TV Comparative proteomics of colon cancer stem cells and differentiated tumor cells identifies BIRC6 as a potential therapeutic target Mol Cell Proteomics 2011 10 12 M111.11353 21788403
- Ren F Wu H Lei Y Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma Mol Cancer 2010 9 81 20403181
- Patel N Chatterjee SK Vrbanac V Rescue of paclitaxel sensitivity by repression of Prohibitin1 in drug-resistant cancer cells Proc Natl Acad Sci U S A 2010 107 6 2503 2508 20133800
- Tilghman SL Townley I Zhong Q Proteomic signatures of acquired letrozole resistance in breast cancer: suppressed estrogen signaling and increased cell motility and invasiveness Mol Cell Proteomics 2013 12 9 2440 2455 23704778
- Zheng S Zhong Q Jiang Q Discovery of a series of thiazole derivatives as novel inhibitors of metastatic cancer cell migration and invasion ACS Med Chem Lett 2013 4 2 191 196 23526571
- Drewes G Chemical proteomics in drug discovery Methods Mol Biol 2012 803 15 21 22065215
- Rix U Superti-Furga G Target profiling of small molecules by chemical proteomics Nat Chem Biol 2009 5 9 616 624 19690537
- Bantscheff M Scholten A Heck AJ Revealing promiscuous drug-target interactions by chemical proteomics Drug Discov Today 2009 14 21–22 1021 1029 19596079
- Dadvar P O’Flaherty M Scholten A Rumpel K Heck AJ A chemical proteomics based enrichment technique targeting the interactome of the PDE5 inhibitor PF-4540124 Mol Biosyst 2009 5 5 472 482 19381362
- Bantscheff M Eberhard D Abraham Y Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors Nat Biotechnol 2007 25 9 1035 1044 17721511
- Trinkle-Mulcahy L Boulon S Lam YW Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes J Cell Biol 2008 183 2 223 239 18936248
- Sutton CW The role of targeted chemical proteomics in pharmacology Br J Pharmacol 2012 166 2 457 475 22074351
- Nomura DK Dix MM Cravatt BF Activity-based protein profiling for biochemical pathway discovery in cancer Nat Rev Cancer 2010 10 9 630 638 20703252
- Gupta A Raina V Geftinib J Cancer Res Ther 2010 6 3 249 254 21119248
- Sanford M Scott LJ Gefitinib: a review of its use in the treatment of locally advanced/metastatic non-small cell lung cancer Drugs 2009 69 16 2303 2328 19852530
- Ranson M Hammond LA Ferry D ZD1839, a selective oral epidermal growth factor receptor-tyrosine kinase inhibitor, is well tolerated and active in patients with solid, malignant tumors: results of a phase I trial J Clin Oncol 2002 20 9 2240 2250 11980995
- Baselga J Rischin D Ranson M Phase I safety, pharmacokinetic, and pharmacodynamic trial of ZD1839, a selective oral epidermal growth factor receptor tyrosine kinase inhibitor, in patients with five selected solid tumor types J Clin Oncol 2002 20 21 4292 4302 12409327
- Gregoire V Hamoir M Chen C Gefitinib plus cisplatin and radiotherapy in previously untreated head and neck squamous cell carcinoma: a phase II, randomized, double-blind, placebo-controlled study Radiother Oncol 2011 100 1 62 69 21821303
- Perez CA Song H Raez LE Phase II study of gefitinib adaptive dose escalation to skin toxicity in recurrent or metastatic squamous cell carcinoma of the head and neck Oral Oncol 2012 48 9 887 892 22513208
- Chen Y Low TY Choong LY Phosphoproteomics identified Endofin, DCBLD2, and KIAA0582 as novel tyrosine phosphorylation targets of EGF signaling and Iressa in human cancer cells Proteomics 2007 7 14 2384 2397 17570516
- Pernas FG Allen CT Winters ME Proteomic signatures of epidermal growth factor receptor and survival signal pathways correspond to gefitinib sensitivity in head and neck cancer Clin Cancer Res 2009 15 7 2361 2372 19318490
- Wu Z Doondeea JB Gholami AM Quantitative chemical proteomics reveals new potential drug targets in head and neck cancer Mol Cell Proteomics 2011 10 12 M111.011635
- Shi H Zhang CJ Chen GY Yao SQ Cell-based proteome profiling of potential dasatinib targets by use of affinity-based probes J Am Chem Soc 2012 134 6 3001 3014 22242683
- Li J Rix U Fang B A chemical and phosphoproteomic characterization of dasatinib action in lung cancer Nat Chem Biol 2010 6 4 291 299 20190765
- Winfield LL Payton-Stewart F Celecoxib and Bcl-2: emerging possibilities for anticancer drug design Future Med Chem 2012 4 3 361 383 22393942
- Kismet K Akay MT Abbasoglu O Ercan A Celecoxib: a potent cyclooxygenase-2 inhibitor in cancer prevention Cancer Detect Prev 2004 28 2 127 142 15068837
- Fosslien E Molecular pathology of cyclooxygenase-2 in neoplasia Ann Clin Lab Sci 2000 30 1 3 21 10678579
- Sareddy GR Geeviman K Ramulu C Babu PP The nonsteroidal anti-inflammatory drug celecoxib suppresses the growth and induces apoptosis of human glioblastoma cells via the NF-κB pathway J Neurooncol 2012 106 1 99 109 21847707
- Takada Y Bhardwaj A Potdar P Aggarwal BB Nonsteroidal anti-inflammatory agents differ in their ability to suppress NF-kappaB activation, inhibition of expression of cyclooxygenase-2 and cyclin D1, and abrogation of tumor cell proliferation Oncogene 2004 23 57 9247 9258 15489888
- Wu T Leng J Han C Demetris AJ The cyclooxygenase-2 inhibitor celecoxib blocks phosphorylation of Akt and induces apoptosis in human cholangiocarcinoma cells Mol Cancer Ther 2004 3 3 299 307 15026550
- Liu DB Hu GY Long GX Qiu H Mei Q Hu GQ Celecoxib induces apoptosis and cell-cycle arrest in nasopharyngeal carcinoma cell lines via inhibition of STAT3 phosphorylation Acta Pharmacol Sin 2012 33 5 682 690 22504904
- Lee EJ Kim SH Kwark YE Kim J Heterogeneous nuclear ribonuclear protein C is increased in the celecoxib-induced growth inhibition of human oral squamous cell carcinoma Exp Mol Med 2006 38 3 203 209 16819278
- Lou J Fatima N Xiao Z Proteomic profiling identifies cyclooxygenase-2-independent global proteomic changes by celecoxib in colorectal cancer cells Cancer Epidemiol Biomarkers Prev 2006 15 9 1598 1606 16985019
- Polakis P Wnt signaling in cancer Cold Spring Harb Perspect Biol 2012 4 5 pii: a008052
- Prosperi JR Goss KH A Wnt-ow of opportunity: targeting the Wnt/beta-catenin pathway in breast cancer Curr Drug Targets 2010 11 9 1074 1088 20545611
- Yao H Ashihara E Maekawa T Targeting the Wnt/β-catenin signaling pathway in human cancers Expert Opin Ther Targets 2011 15 7 873 887 21486121
- Huang SM Mishina YM Liu S Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling Nature 2009 461 7264 614 620 19759537
- Seimiya H The telomeric PARP, tankyrases, as targets for cancer therapy Br J Cancer 2006 94 3 341 345 16421589
- Karlberg T Markova N Johansson I Structural basis for the interaction between tankyrase-2 and a potent Wnt-signaling inhibitor J Med Chem 2010 53 14 5352 5355 20565110
- Gills JJ Jeffery EH Matusheski NV Moon RC Lantvit DD Pezzuto JM Sulforaphane prevents mouse skin tumorigenesis during the stage of promotion Cancer Lett 2006 236 1 72 79 15993536
- Myzak MC Dashwood RH Chemoprotection by sulforaphane: keep one eye beyond Keap1 Cancer Lett 2006 233 2 208 218 16520150
- Fimognari C Hrelia P Sulforaphane as a promising molecule for fighting cancer Mutat Res 2007 635 2–3 90 104 17134937
- Clarke JD Dashwood RH Ho E Multi-targeted prevention of cancer by sulforaphane Cancer Lett 2008 269 2 291 304 18504070
- Mastrangelo L Cassidy A Mulholland F Wang W Bao Y Serotonin receptors, novel targets of sulforaphane identified by proteomic analysis in Caco-2 cells Cancer Res 2008 68 13 5487 5491 18593952
- Lee CH Jeong SJ Yun SM Down-regulation of phosphoglucomutase 3 mediates sulforaphane-induced cell death in LNCaP prostate cancer cells Proteome Sci 2010 8 67 21159204
- Borrebaeck CA Viewpoints in clinical proteomics: when will proteomics deliver clinically useful information? Proteomics Clin Appl 2012 6 7–8 343 345 22696166
- Boschetti E Chung MC Righetti PG “The quest for biomarkers”: are we on the right technical track? Proteomics Clin Appl 2012 6 1–2 22 41 22213582
- Dasilva N Díez P Matarraz S Biomarker discovery by novel sensors based on nanoproteomics approaches Sensors (Basel) 2012 12 2 2284 2308 22438764
- Dubois E Fertin M Burdese J Amouyel P Bauters C Pinet F Cardiovascular proteomics: translational studies to develop novel biomarkers in heart failure and left ventricular remodeling Proteomics Clin Appl 2011 5 1–2 57 66 21246740
- Geho DH Petricoin EF Liotta LA Blasting into the microworld of tissue proteomics: a new window on cancer Clin Cancer Res 2004 10 3 825 827 14871957
- Rho JH Roehrl MH Wang JY Tissue proteomics reveals differential and compartment-specific expression of the homologs transgelin and transgelin-2 in lung adenocarcinoma and its stroma J Proteome Res 2009 8 12 5610 5618 19848416
- Lehtiö J De Petris L Lung cancer proteomics, clinical and technological considerations J Proteomics 2010 73 10 1851 1863 20685322
- Peng XC Gong FM Zhao YW Comparative proteomic approach identifies PKM2 and cofilin-1 as potential diagnostic, prognostic and therapeutic targets for pulmonary adenocarcinoma PLoS One 2011 6 11 e27309 22087286
- Carretero J Shimamura T Rikova K Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors Cancer Cell 2010 17 6 547 559 20541700
- Tian T Hao J Xu A Determination of metastasis-associated proteins in non-small cell lung cancer by comparative proteomic analysis Cancer Sci 2007 98 8 1265 1274 17537172
- Yao H Zhang Z Xiao Z Identification of metastasis associated proteins in human lung squamous carcinoma using two-dimensional difference gel electrophoresis and laser capture microdissection Lung Cancer 2009 65 1 41 48 19058872
- Vander Heiden MG Christofk HR Schuman E Identification of small molecule inhibitors of pyruvate kinase M2 Biochem Pharmacol 2010 79 8 1118 1124 20005212
- Eiseler T Döppler H Yan IK Kitatani K Mizuno K Storz P Protein kinase D1 regulates cofilin-mediated F-actin reorganization and cell motility through slingshot Nat Cell Biol 2009 11 5 545 556 19329994
- Nakashima S Matsuda H Kurume A Cucurbitacin E as a new inhibitor of cofilin phosphorylation in human leukemia U937 cells Bioorg Med Chem Lett 2010 20 9 2994 2997 20347305
- Goo YA Goodlett DR Advances in proteomic prostate cancer biomarker discovery J Proteomics 2010 73 10 1839 1850 20398807
- Larkin SE Zeidan B Taylor MG Proteomics in prostate cancer biomarker discovery Expert Rev Proteomics 2010 7 1 93 102 20121479
- Ummanni R Mundt F Pospisil H Identification of clinically relevant protein targets in prostate cancer with 2D-DIGE coupled mass spectrometry and systems biology network platform PLoS One 2011 6 2 e16833 21347291
- Azimzadeh O Barjaktarovic Z Aubele M Formalin-fixed paraffin-embedded (FFPE) proteome analysis using gel-free and gel-based proteomics J Proteome Res 2010 9 9 4710 4720 20604508
- Tanca A Pagnozzi D Addis MF Setting proteins free: progresses and achievements in proteomics of formalin-fixed, paraffin-embedded tissues Proteomics Clin Appl 2012 6 1–2 7 21 22213597
- Ralton LD Murray GI The use of formalin fixed wax embedded tissue for proteomic analysis J Clin Pathol 2011 64 4 297 302 21285242
- Hara A Sakai N Yamada H Yoshimi N Tanaka T Mori H Immunoblot analysis of the placental form of glutathione S-transferase in protein extracted from paraffin-embedded human glioma tissue J Cancer Res Clin Oncol 1993 119 8 493 496 8509440
- Layfield R Bailey K Dineen R Application of formalin fixation to the purification of amyloid proteins Anal Biochem 1997 253 1 142 144 9356158
- Layfield R Bailey K Lowe J Allibone R Mayer RJ Landon M Extraction and protein sequencing of immunoglobulin light chain from formalin-fixed cerebrovascular amyloid deposits J Pathol 1996 180 4 455 459 9014869
- Shi SR Liu C Balgley BM Lee C Taylor CR Protein extraction from formalin-fixed, paraffin-embedded tissue sections: quality evaluation by mass spectrometry J Histochem Cytochem 2006 54 6 739 743 16399996
- Shi SR Shi Y Taylor CR Antigen retrieval immunohistochemistry: review and future prospects in research and diagnosis over two decades J Histochem Cytochem 2011 59 1 13 32 21339172
- Chu WS Liang Q Liu J A nondestructive molecule extraction method allowing morphological and molecular analyses using a single tissue section Lab Invest 2005 85 11 1416 1428 16127423
- Sprung RW Brock JW Tanksley JP Equivalence of protein inventories obtained from formalin-fixed paraffin-embedded and frozen tissue in multidimensional liquid chromatography-tandem mass spectrometry shotgun proteomic analysis Mol Cell Proteomics 2009 8 8 1988 1998 19467989
- Ikeda K Monden T Kanoh T Extraction and analysis of diagnostically useful proteins from formalin-fixed, paraffin-embedded tissue sections J Histochem Cytochem 1998 46 3 397 403 9487122
- Prieto DA Hood BL Darfler MM Liquid tissue: proteomic profiling of formalin-fixed tissues Biotechniques 2005 Suppl:32 35 16528915
- Palmer-Toy DE Krastins B Sarracino DA Nadol JBJr Merchant SN Efficient method for the proteomic analysis of fixed and embedded tissues J Proteome Res 2005 4 6 2404 2411 16335994
- Crockett DK Lin Z Vaughn CP Lim MS Elenitoba-Johnson KS Identification of proteins from formalin-fixed paraffin-embedded cells by LC-MS/MS Lab Invest 2005 85 11 1405 1415 16155593
- Hwang SI Thumar J Lundgren DH Direct cancer tissue proteomics: a method to identify candidate cancer biomarkers from formalin-fixed paraffin-embedded archival tissues Oncogene 2007 26 1 65 76 16799640
- Rahimi F Shepherd CE Halliday GM Geczy CL Raftery MJ Antigen-epitope retrieval to facilitate proteomic analysis of formalin-fixed archival brain tissue Anal Chem 2006 78 20 7216 7221 17037924
- Knight WA Livingston RB Gregory EJ McGuire WL Estrogen receptor as an independent prognostic factor for early recurrence in breast cancer Cancer Res 1977 37 12 4669 4671 922747
- Mason BH Holdaway IM Mullins PR Yee LH Kay RG Progesterone and estrogen receptors as prognostic variables in breast cancer Cancer Res 1983 43 6 2985 2990 6850609
- Chevallier B Heintzmann F Mosseri V Prognostic value of estrogen and progesterone receptors in operable breast cancer. Results of a univariate and multivariate analysis Cancer 1988 62 12 2517 2524 3191451
- Henry JA Nicholson S Farndon JR Westley BR May FE Measurement of oestrogen receptor mRNA levels in human breast tumours Br J Cancer 1988 58 5 600 605 2851309
- Rezaul K Thumar JK Lundgren DH Differential protein expression profiles in estrogen receptor-positive and -negative breast cancer tissues using label-free quantitative proteomics Genes Cancer 2010 1 3 251 271 21779449
- Cabezón T Gromova I Gromov P Proteomic profiling of triple-negative breast carcinomas in combination with a three-tier orthogonal technology approach identifies Mage-A4 as potential therapeutic target in estrogen receptor negative breast cancer Mol Cell Proteomics 2013 12 2 381 394 23172894