Abstract
Purpose
Brown adipose tissue (BAT) can rapidly generate heat and improve energy metabolism. Circular RNAs (circRNAs) are cellular endogenous non-coding RNAs, which can regulate the development and progress of different diseases. However, the role of circRNAs in human BAT is not fully understood. Here, we analyzed the differentially expressed circRNAs (DECs) in human BAT, as well as in white adipose tissue (WAT), and identified new biomarkers of BAT.
Patients and Methods
Three human BAT and three human subcutaneous WAT samples were selected, and circRNA microarray was performed. Additionally, quantitative real-time polymerase chain reaction (qRT-PCR) was applied to determine the expression of six circRNAs. Finally, the functional analysis was performed by bioinformatics.
Results
Compared to WAT, 152 upregulated circRNAs and 201 downregulated circRNAs were identified in BAT. The DECs were further subjected to GO and KEGG enrichment analysis. Several circRNAs, for example, hsa_circ_0006168, hsa_circ_26337 and hsa_circ_0007507 were found upregulated and hsa_circ_0030162 was found downregulated in human BAT compared to WAT.
Conclusion
This study profiles the circRNA expression in human BAT and WAT, and suggests hsa_circ_0006168, hsa_circ_26337, hsa_circ_0007507, and hsa_circ_0030162 as novel biomarkers for human BAT.
Introduction
The excessive accumulation of adipose tissue is one of the leading factors of obesity.Citation1,Citation2 Adipose tissues are typed as white adipose tissue (WAT), brown adipose tissue (BAT) and beige adipose tissue.Citation3 The primary function of WAT is storing triglyceride, while BAT, in contrast to WAT, dissipates the energy of chemical bindings through uncoupling protein 1 (UCP1) that results in increased fatty acid oxidation and thermogenesis.Citation4,Citation5 Biologically active BAT can promote thermogenesis through UCP1, and activation of BAT and induction of WAT browning accelerates glycolipid uptake, and is essential for improving lipid metabolism and insulin resistance.Citation6 However, beige adipose tissue has the characteristics of both WAT and BAT, which can store triglyceride and promote thermogenesis after cold stimulation.Citation7,Citation8 BAT has recently gained importance in metabolic research, partially attributed to its unique metabolic function.Citation9–11 Thus, new BAT biomarkers are required to be explored for clinical practices.
Circular RNAs (circRNAs), a special member of noncoding RNAs, are closed in structure, and formed by covalent bonding.Citation12–14 They are widely expressed in mammals.Citation14–16 Numerous studies indicated that circRNAs play a critical role in the development of various diseases.Citation17–21 Recent reports indicate that circRNAs are differentially expressed in mice BAT and WAT,Citation22,Citation23 but the pattern of differentially expressed circRNAs in human BAT and WAT remains unclear.
In the present study, we analyzed the expression profiles of circRNA by using circRNA microarray to explore the differentially expressed circRNAs (DECs) in human BAT and WAT. Subsequently, six differentially expressed circRNAs were chosen for verifying the expression through qRT-PCR. Among them, four circRNAs were validated in human BAT and the biological functions were predicted by further bioinformatics analysis. Our study is the first comprehensive analysis of human BAT circRNA expression profiles, and identification of new biomarkers of BAT provides new strategies for the prevention and treatment of obesity and related disease.
Materials and Methods
Research Subjects
In this study, 30 patients with clavicle fractures and 3 patients with cholecystectomy were included. All patients were admitted in the Department of Orthopaedic Surgery and Department of Hepatobiliary Surgery of the Third Xiangya Hospital of Central South University from November 2021 to June 2022. All patients were excluded from diabetes, acute infection, tumor, and smoking. The adipose tissue samples were obtained from the supraclavicular fossa of 30 patients with clavicle fractures, but only 3 cases of BAT were confirmed with H&E staining and UCP1 expression. WAT samples were obtained from abdominal subcutaneous tissues of three patients with cholecystectomy surgery; those patients were of same gender, age, and weight with three BAT group patients.
The design of this research work was approved by the Ethics Review Committee of the Third Xiangya Hospital of Central South University, Changsha, China, and the research experiments were performed according to the guidelines outlined in the Declaration of Helsinki. All individual patients provided written informed consent.
H&E Staining
The paraffin-embedded adipose tissue was cut into small sections, dewaxed twice through xylene, and sections were sequentially soaked in alcohol at different concentrations followed by washing with distilled water. The sections were then stained with hematoxylin (Servicebio, Wuhan, China), washed in distilled water, dehydrated with 0.5% hydrochloric acid alcohol. Next, the sections were stained in an eosin staining solution (Servicebio, Wuhan, China), and dried after washing with distilled water. Lastly, neutral resin (Servicebio, Wuhan, China) was used to seal the sections, and photos were taken under a microscope (Leica DM IL LED Fluo, Germany).
Total RNA Extraction with RNA Labeling and Hybridization
Total RNA materials were obtained from human BAT and WAT samples by using trizol (Invitrogen, Carlsbad, CA, USA). The total RNA was then treated with Rnase R (Epicentre, Madison, WI, USA), and the linear RNA was removed for enriching circRNAs. The circRNAs were amplified by random primer method (Arraystar Super RNA Labeling Kit, Arraystar, Rockville, MD, USA), and then transcribed to fluorescent cRNA. Subsequently, the labeled fluorescent cRNA was recovered (Qiagen, Hilden, Germany), and then subjected to activity and concentration measurement by using a spectrophotometer (NanoDrop ND-1000, NanoDrop Technologies, Wilmington, DE, USA). Next, 5-µL 10× blocker and 1-µL 25× fragmentation buffer was added to each labeled cRNA to make them fragmented and the mixture was kept at 60°C for 30 min, followed by the addition of 25-µL 2× hybridization buffer. Next, 50 μL hybridization solution was used to assemble onto circRNA expression chip slides. The slides were then incubated for 17 hours at 65°C. Finally, after cleaning and fixing the hybridized arrays, the results were produced by using Agilent scanner G2505C (Agilent Technologies, Santa Clara, CA, USA).
CircRNA Microarray Analysis
Raw data were extracted through Agilent Feature Extraction software (Agilent Technologies, Santa Clara, CA, USA), and then uploaded to the R software limma package for quantile normalization as well as subsequent data processing. By low-intensity filtering, we found at least three out of six circRNAs for further analyses. Volcano plot and fold change cutoff were combined to identify DEG with statistical significance between two groups or samples.
Gene Ontology (GO) enrichment was conducted by using the GO seq R package. Encyclopedia of Genes and Genomes (KEGG) pathway enrichment was achieved by DAVID online tool (https://david.ncifcrf.gov/).
QRT-PCR Amplification of Partial circRNAs
Human BAT and WAT were analyzed based on raw data and validated using qRT-PCR. cDNA was reverse transcribed by Revert Aid TMM-MuLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA). qRT-PCR was performed by using a thermocycler (Applied Biosystems, New York, NY, USA). The primer sequences were designed by circPrimer1.2 (http://www.bioinf.com.cn/) and are listed in Supplementary Table 1.
The circRNA–microRNA–mRNA Network Prediction
For exploring the biological function of circRNAs, the circRNA–microRNA binding sites were predicted by online software miRanda and TargetScan. The DECs within all the comparisons were marked with circRNA–microRNA interaction information. The miRNA and mRNA interaction prediction was performed by using the software, DIANA TOOLS. And the circRNA–microRNA–mRNA interaction was constructed and visualized by Cytoscape 2.8.2.Citation15
Statistics Analysis
The data analyses between two groups were performed with paired t‐test (GraphPad Prism 8.0, GraphPad, CA, USA), where P<0.05 was considered statistically significant.
Results
Differential Expression Profile of circRNAs in BAT and WAT
H&E staining was used to verify phenotypic characters of patients’ sample from which BAT or WAT obtained. WAT had a larger cell size and uni-locular lipid droplets, while in BAT there was a smaller cell size and multi-locular lipid droplet (). In addition, UCP1, the molecular marker of BAT, was significantly upregulated in BAT compared to WAT (). Next, DECs between BAT and WAT were detected with a human circRNA microarray. The box plot showed that the Log2 ratio distribution was comparable in all of the samples (). Thus, the results suggested that circRNAs in BAT had an expression pattern that varied largely from WAT. The hierarchical clustering results indicated that the circRNAs expression profiles of BAT were apparently different from that of WAT (). The Scatter plot also showed a clear discrepancy in circRNAs expression between BAT and WAT (). The volcano plot showed that DECs between BAT and WAT have statistical significance (multiplicity >2.0, P<0.05) (). Our analyses suggested that circRNA expression patterns are different in BAT compared to WAT.
Figure 1 H&E staining and UCP1 expression in BAT and WAT. (A) BAT and WAT were stained with H&E, scale bar 200μm. (B) UCP1 mRNA expression of BAT and WAT. n=3, **p<0.01.
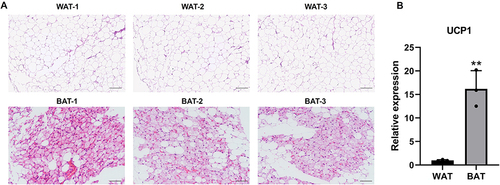
Figure 2 DECs in human BAT and WAT. (A) The box plot displayed the normalized intensity distribution of all data. (B) Hierarchical clustering maps of DECs were shown in BAT and WAT. Red represents relatively high expression; green represents relatively low expression. (C) Scatter plots were used to assess changes in circRNAs expression between BAT and WAT. CircRNAs above the top green line and below the bottom green line indicate more than a 1.5-fold variation in circRNAs between the two comparison samples. (D) The volcano plot showed statistically significant DECs between BAT and WAT (fold change >2.0, P<0.05). (E) Classification and distribution of DECs in human chromosomes.
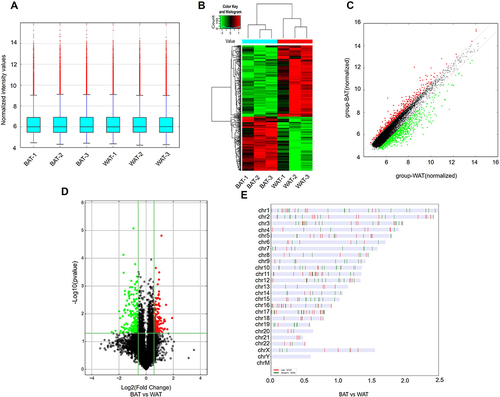
Based on the above results, we found that there were 353 DECs in BAT compared to WAT, of which 152 circRNAs were upregulated and 201 circRNAs were downregulated. The leading 10 upregulated and downregulated circRNAs are shown in and , respectively. The categories and distribution of these DECs in human chromosomes were identified indicating that most of these circRNAs are located on chromosome 1–4 ().
Table 1 Upregulated circRNAs in Human BAT Vs WAT
Table 2 Downregulated circRNAs in Human BAT Vs WAT
Bioinformatic Analysis of DECs
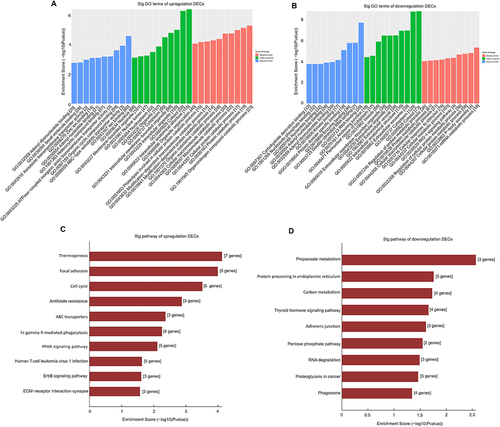
We performed GO and KEGG analyses for DECs according to their host genes. In upregulated circRNAs, the GO analysis indicated that DECs in BAT were linked to organonitrogen compound catabolic process, intracellular anatomical structure, and ion binding, and so on (). In the downregulated circRNAs, DECs of GO analysis were enriched in the mRNA metabolic process, cytoplasm and RNA binding, and so on (). In upregulated circRNAs, KEGG analysis displayed that the most noticeable enriched pathways were thermogenesis, focal adhesion, and cell cycle (). However, in downregulated circRNAs, the most significantly enriched pathways were found propanoate metabolism, protein processing in the endoplasmic reticulum, and carbon metabolism ().
Validation of Expression of DECs
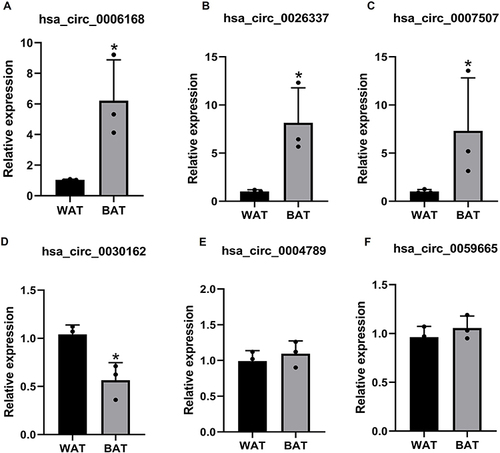
The four circRNAs with most upregulated expression and two circRNAs most downregulated expression according to fold change were verified with qRT-PCR analysis, and the partial circRNAs did not show statistical difference in circRNA microarray result. Our results showed that hsa_circ_0006168, hsa_circ_26337, and hsa_circ_0007507 expressions are significantly elevated in BAT compared to WAT. In contrast, the expression of hsa_circ_0030162 was significantly declined, which is consistent with circRNAs microarray expression profiles (). However, the level of hsa_circ_0004789 and hsa_circ_0059662 had no noticeable difference between the two groups ( and ). So, we propose that hsa_circ_0006168, hsa_circ_26337, hsa_circ_0007507, and hsa_circ_0030162 are novel biomarkers for human BAT.
Prediction of circRNAs Targeting microRNA–mRNA Network
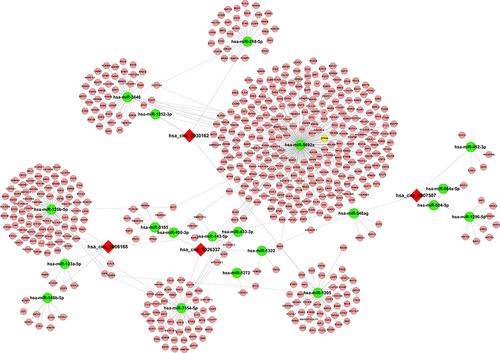
To further evaluate the potential function of interested circRNAs, we predicted the potential circRNA-microRNA network by TargetScan and miRanda. According to the highest prediction score, our results showed five microRNAs interacting with hsa_circ_0006168, hsa_circ_26337, hsa_circ_0007507, and hsa_circ_0030162, respectively. For instance, hsa-miR-100-3p, hsa-miR-125b-5p, hsa-miR-133a-5p, hsa-miR-143-5p and hsa-miR-148b-5p might be the target microRNAs of hsa_circ_0006168, while hsa-miR-452-3p, hsa-miR-548ag, hsa-miR-1296-5p, hsa-miR-504-5p and hsa-miR-664a-5p were predicted as the targets of hsa_circ_0007507. The interaction network of circRNA–microRNA–mRNA is summarized in .
Figure 5 Prediction of the circRNA–microRNA–mRNA interaction network. The target microRNAs of hsa_circ_0006168 (A), hsa_circ_0026337 (B), hsa_circ_000750 (C), and hsa_circ_0030162 (D) were predicted by Arraystar’s home-made software and target mRNA of microRNA was predicted by DIANA TOOLS software. circRNA–microRNA–mRNA interaction network was visualized and constructed by Cytoscape 2.8.2.
Discussion
Adipose tissues are important targets for treating metabolic disorders, like obesity, and BAT and WAT play different roles in regulating metabolism.Citation24,Citation25 CircRNAs were reported to be involved in the development and progression of various metabolic diseases.Citation26,Citation27 Xu et al found that overexpression of circRNA-CDR1as is associated with diabetes improved insulin levels in mice.Citation28 Li et al reported that circScd1 overexpression promotes steatosis in non-alcoholic fatty liver.Citation29 Previous reports also showed that circRNAs are abundantly expressed in BAT and WAT of mice,Citation22,Citation23 however, there is no clear report on circRNA expression profiles in human BAT and WAT. So, here we aimed to explore the DECs between BAT and WAT, and attempted to identify some novel biomarkers for metabolic diseases.
In GO and KEGG analysis of DECs, and we tried to predict the functions of circRNAs in human BAT. The results indicated that DECs are associated with cellular proteolytic metabolic processes. The most significantly enriched pathways include thermogenesis focal adhesion cell cycle, and propanoate metabolism, etc, in particular, some circRNAs are associated with the thermogenesis pathway, which is consistent with the molecular characteristics of BAT. Moreover, the cell cycle is a critical pathway to participate in the adipogenesis of BAT.Citation30
In addition, we verified six DECs by qRT-PCR, and found that hsa_circ_0006168, hsa_circ_26337, hsa_circ_0007507, and hsa_circ_0030162 expression patterns are consistent with the circRNA microarray data. hsa_circ_0006168 has been reported to participate in regulating the proliferation and invasion of glioblastoma and esophageal cancer cells.Citation31–33 In addition, hsa_circRNA_0007507 and its host gene, RASA1, are upregulated in gastric cancer.Citation34
Usually circRNAs act as a sponge for microRNAs.Citation14,Citation35,Citation36 Thus, we predicted the potential association network of circRNAs with its binding target microRNAs and mRNAs. For instance, miR-125b-5p, miR-133a-5p, and miR-143-5p were predicted to interact with hsa_circ_0006168. Interestingly, miR-125b-5p is negatively regulated by UCP1 expression and BAT formation in humans and mice. Furthermore, miR-125b inhibited Brite adipose formation.Citation37 Knockout of miR-133a improved BAT thermogenic gene expression including PRDM16, UCP1 and CIDEA, and promoted BAT adipogenesis.Citation38–40 Knockout miR-143 elevated thermogenesis and lipolysis in BAT and decrease the BAT formation.Citation41 miR-143 was also reported to be downregulated in BAT than WAT in case of both of pre- and mature adipocytes.Citation42 Although we did not find studies on other’s predictions on microRNAs of hsa_circ_26337, hsa_circ_0007507, and hsa_circ_003016, we speculate that hsa_circ_0006168 most likely plays a vital role in BAT function.
This study had some limitations too. Firstly, the sample size was small; although we collected 30 cases of adipose tissue from the patient’s supraclavicular fossa, only 3 cases of BAT were verified by H&E staining and UCP1 expression. Secondly, only a small number of circRNAs were validated. So further investigations are required into these perspectives in future research works.
Conclusion
In this study, by using circRNA microarray, we identified several DECs including hsa_circ_0006168, hsa_circ_26337, hsa_circ_0007507, and hsa_circ_0030162 between human BAT and WAT. Our findings indicated that these circRNAs biomarkers are associated with adipose tissue formation, as well as lipolysis, and they are identified as potential therapeutic targets for treating obesity and related diseases. The regulatory mechanisms of these circRNAs will be further elucidated in future studies.
Abbreviations
BAT, brown adipose tissue; WAT, white adipose tissue; circRNAs, circular RNAs; DECs, differentially expressed circRNAs; qRT-PCR, quantitative real-time polymerase chain reaction; UCP1, uncoupling protein 1; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Disclosure
The authors report that there are no conflicts of interest regarding this work.
Additional information
Funding
References
- Navarro-Ruiz MDC, López-Alcalá J, Díaz-Ruiz A, et al. Understanding the adipose tissue acetylome in obesity and insulin resistance. Transl Res. 2022;246:15–32. doi:10.1016/j.trsl.2022.02.008
- González-Domínguez Á, Visiedo-García FM, Domínguez-Riscart J, et al. Iron metabolism in obesity and metabolic syndrome. Int J Mol Sci. 2020;21(15):5529. doi:10.3390/ijms21155529
- Jiang S, Lin J, Zhang Q, et al. The fates of different types of adipose tissue after transplantation in mice. FASEB J. 2022;36(9):e22510. doi:10.1096/fj.202200408R
- Marlatt KL, Ravussin E. Brown adipose tissue: an update on recent findings. Curr Obes Rep. 2017;6(4):389–396. doi:10.1007/s13679-017-0283-6
- Čater M, Križančić Bombek L. Protective role of mitochondrial uncoupling proteins against age-related oxidative stress in type 2 diabetes mellitus. Antioxidants. 2022;11(8):1473. doi:10.3390/antiox11081473
- Cheng L, Wang J, Dai H, et al. Brown and beige adipose tissue: a novel therapeutic strategy for obesity and type 2 diabetes mellitus. Adipocyte. 2021;10(1):48–65. doi:10.1080/21623945.2020.1870060
- Ishibashi J, Seale P. Medicine. Beige can be slimming. Science. 2010;328(5982):1113–1114. doi:10.1126/science.1190816
- Giralt M, Villarroya F. White, brown, beige/brite: different adipose cells for different functions? Endocrinology. 2013;154(9):2992–3000. doi:10.1210/en.2013-1403
- Zhang Q, Ye R, Zhang YY, et al. Brown adipose tissue and novel management strategies for polycystic ovary syndrome therapy. Front Endocrinol. 2022;13:847249. doi:10.3389/fendo.2022.847249
- Virtanen KA, Lidell ME, Orava J, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360(15):1518–1525. doi:10.1056/NEJMoa0808949
- Oelkrug R, Polymeropoulos ET, Jastroch M, et al. Brown adipose tissue: physiological function and evolutionary significance. J Comp Physiol B. 2015;185(6):587–606. doi:10.1007/s00360-015-0907-7
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
- Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi:10.1016/j.canlet.2015.06.003
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi:10.1016/j.molcel.2014.08.019
- Huang C, Shan G. What happens at or after transcription: insights into circRNA biogenesis and function. Transcription. 2015;6(4):61–64. doi:10.1080/21541264.2015.1071301
- Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi:10.1016/j.omtn.2019.02.005
- Li M, Duan L, Li Y, et al. Long noncoding RNA/circular noncoding RNA-miRNA-mRNA axes in cardiovascular diseases. Life Sci. 2019;233:116440. doi:10.1016/j.lfs.2019.04.066
- Panda AC, Grammatikakis I, Kim KM, et al. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res. 2017;45(7):4021–4035. doi:10.1093/nar/gkw1201
- Zhou R, Wu Y, Wang W, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–142. doi:10.1016/j.canlet.2018.03.035
- Lai S, Du K, Shi Y, et al. Long non-coding RNAs in brown adipose tissue. Diabetes Metab Syndr Obes. 2020;13:3193–3204. doi:10.2147/DMSO.S264830
- Liu K, Liu X, Deng Y, et al. CircRNA-mediated regulation of brown adipose tissue adipogenesis. Front Nutr. 2022;9:926024. doi:10.3389/fnut.2022.926024
- Zhang P, Sheng M, Du C, et al. Assessment of CircRNA expression profiles and potential functions in brown adipogenesis. Front Genet. 2021;12:769690. doi:10.3389/fgene.2021.769690
- Kawai T, Autieri MV, Scalia R. Adipose tissue inflammation and metabolic dysfunction in obesity. Am J Physiol Cell Physiol. 2021;320(3):C375–C391. doi:10.1152/ajpcell.00379.2020
- Ronti T, Lupattelli G, Mannarino E. The endocrine function of adipose tissue: an update. Clin Endocrinol. 2006;64(4):355–365. doi:10.1111/j.1365-2265.2006.02474.x
- Wang T, Pan W, Hu J, et al. Circular RNAs in metabolic diseases. Adv Exp Med Biol. 2018;1087:275–285. doi:10.1007/978-981-13-1426-1_22
- Zaiou M. Circular RNAs as potential biomarkers and therapeutic targets for metabolic diseases. Adv Exp Med Biol. 2019;1134:177–191. doi:10.1007/978-3-030-12668-1_10
- Xu H, Guo S, Li W, et al. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5(1):12453. doi:10.1038/srep12453
- Li P, Shan K, Liu Y, et al. CircScd1 promotes fatty liver disease via the janus kinase 2/signal transducer and activator of transcription 5 pathway. Dig Dis Sci. 2019;64(1):113–122. doi:10.1007/s10620-018-5290-2
- Lopez-Mejia IC, Castillo-Armengol J, Lagarrigue S, et al. Role of cell cycle regulators in adipose tissue and whole body energy homeostasis. Cell Mol Life Sci. 2018;75(6):975–987. doi:10.1007/s00018-017-2668-9
- Shi Y, Guo Z, Fang N, et al. hsa_circ_0006168 sponges miR-100 and regulates mTOR to promote the proliferation, migration and invasion of esophageal squamous cell carcinoma. Biomed Pharmacother. 2019;117:109151. doi:10.1016/j.biopha.2019.109151
- Xie ZF, Li HT, Xie SH, et al. Circular RNA hsa_circ_0006168 contributes to cell proliferation, migration and invasion in esophageal cancer by regulating miR-384/RBBP7 axis via activation of S6K/S6 pathway. Eur Rev Med Pharmacol Sci. 2020;24(1):151–163. doi:10.26355/eurrev_202001_19906
- Wang T, Mao P, Feng Y, et al. Blocking hsa_circ_0006168 suppresses cell proliferation and motility of human glioblastoma cells by regulating hsa_circ_0006168/miR-628-5p/IGF1R ceRNA axis. Cell Cycle. 2021;20(12):1181–1194. doi:10.1080/15384101.2021.1930357
- Zhang W, Zheng M, Kong S, et al. Circular RNA hsa_circ_0007507 may serve as a biomarker for the diagnosis and prognosis of gastric cancer. Front Oncol. 2021;11:699625. doi:10.3389/fonc.2021.699625
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. doi:10.1038/nrm.2015.32
- Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi:10.1016/j.molcel.2018.06.034
- Giroud M, Pisani DF, Karbiener M, et al. miR-125b affects mitochondrial biogenesis and impairs brite adipocyte formation and function. Mol Metab. 2016;5(8):615–625. doi:10.1016/j.molmet.2016.06.005
- Liu W, Bi P, Shan T, et al. miR-133a regulates adipocyte browning in vivo. PLoS Genet. 2013;9(7):e1003626. doi:10.1371/journal.pgen.1003626
- Liu W, Kuang S. miR-133 links to energy balance through targeting Prdm16. J Mol Cell Biol. 2013;5(6):432–434. doi:10.1093/jmcb/mjt036
- Trajkovski M, Ahmed K, Esau CC, et al. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat Cell Biol. 2012;14(12):1330–1335. doi:10.1038/ncb2612
- Liu J, Liu J, Zeng D, et al. miR-143-null is against diet-induced obesity by promoting BAT thermogenesis and inhibiting WAT adipogenesis. Int J Mol Sci. 2022;23(21):13058. doi:10.3390/ijms232113058
- Walden TB, Timmons JA, Keller P, et al. Distinct expression of muscle-specific microRNAs (myomirs) in brown adipocytes. J Cell Physiol. 2009;218(2):444–449. doi:10.1002/jcp.21621