Abstract
Purpose
Streptococcus pneumoniae (S. pneumoniae) is the etiology of severe and life-threatening infections in children less than 5 years old. Though pneumococcal conjugate vaccines (PCVs) are effective in the prevention of pneumococcal infections, yet they are not included in the National Immunization Program in Egypt pending the identification of pathogenic serotypes. As S. pneumoniae colonization of the pharynx predisposes to pneumonia and invasive pneumococcal disease (IPD) caused by the colonizing serotypes, identification of the nasopharyngeal (NP) serotypes can be a surrogate to the invasive serotypes. In this study, we aimed to 1. Identify the serotypes and antimicrobial susceptibility testing (AST) of Streptococcus pneumoniae colonizing the nasopharynx of Egyptian children younger than 5 years in two successive winter seasons. 2. Correlate the identified serotypes with vaccine coverage of the 13-valent conjugate pneumococcal vaccines (PCV13). 3. Compare the serotypes and AST of S. pneumoniae from NP to those of IPD that were routinely identified in our clinical laboratory during the study period.
Materials and Methods
The study was conducted in two successive winter seasons (December 2015–March 2016; December 2016–March 2017). We enrolled 334 children, aged 6 months to 5 years, attending the outpatient general clinics of Cairo University Children Hospital, excluding those with fever, signs of infection, history of antibiotic intake or hospitalization in the preceding month. We tested NP swabs for S. pneumoniae by culture and real-time PCR. Serotyping was performed by sequential multiplex PCR for all positive samples. AST was done to S. pneumoniae isolates by Vitek-2™ (BioMérieux, Marcy-L’Etoile, France). We included routinely detected S. pneumoniae from sterile body sites during the study period, and identified their serotypes and AST.
Results
PCR was positive for pneumococci in 217 out of 334 pharyngeal swabs (65%), including 186 typable samples. The most common serotypes were serotypes 1, 6ABC, 19 F, 5 and 18ABC. By culture, we isolated only 110 out of 334 pharyngeal swabs (32.9%). The theoretical coverage of the PCV13 vaccine for the detected serotypes was 77.4%. The AST of NP isolates revealed low susceptibility rates to all antimicrobials except for vancomycin, linezolid, levofloxacin and clindamycin. During the study period, we identified 40 IPD; 21 identified by PCR and 19 by culture. The commonest pneumococcal serotypes were 1, 18ABC, 6ABC and 5. The PCV13 coverage was 75%. By Vitek-2, the isolates showed 100%, 100%, 94.7%, 89.5%, 84.2%, 84.2% and 78.9% susceptibility to vancomycin, linezolid, clindamycin, levofloxacin, penicillin, cefotaxim and erythromycin, respectively.
Conclusion
Based on the serotype vaccine coverage and the emerging antimicrobial resistance of S. pneumoniae, PCVs will be valuable to Egyptian children.
Introduction
Streptococcus pneumoniae (S. pneumoniae) is one of the most common pathogens causing a wide range of infections. Pneumococcal infections are prevalent in children, with adverse clinical and economic burden, particularly invasive pneumococcal diseases (IPD) and when infection is caused by resistant isolates.Citation1
Though the pneumococcal conjugate vaccines (PCVs) have proved to be effective, many middle-income countries are implementing PCVs at a slower rate than donor-funded low-income countries and wealthier developed countries.Citation2
In Egypt, PCVs have not yet been included in the national children vaccination program as the availing data on pneumococcal serotypes are limited. In the winter season of 2013–2014, a study from Egypt detected nasopharyngeal (NP) carriage of S. pneumoniae in 56.5% of children less than 5 years; and the coverage of the PCV13 vaccine for the detected serotypes was 72.4%.Citation3 However, as the NP pneumococcal serotypes show temporal variations in serotypes and antimicrobial resistance,Citation4 more studies are deemed necessary. In this study, we aimed to:
Identify the serotypes (ST) and antimicrobial susceptibility testing (AST) of S. pneumoniae colonizing the nasopharynx of Egyptian children younger than 5 years in two successive winter seasons, and detect the temporal changes in serotypes in comparison to a recently published report.Citation3
Correlate the identified serotypes with vaccine coverage of PCV13.
Compare the NP colonization of S. pneumoniae serotypes and AST to the serotypes and AST of S. pneumoniae routinely identified from IPD in our clinical laboratory during the study period.
Materials and Methods
Ethical approval was obtained from the Cairo University, Faculty of Medicine REC, that works in accordance with the Declaration of Helsinki under the number R-11-2018. Informed consent was obtained in Arabic language from the guardian.
Subjects
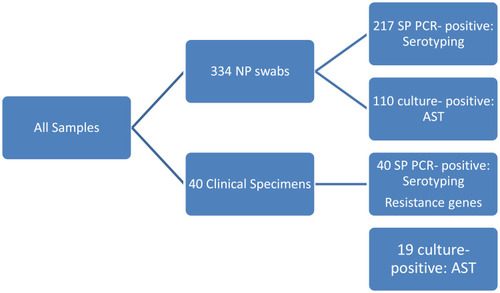
This study was conducted in two successive winter seasons (December 2015 –March 2016; December 2016-March 2017). We enrolled 334 children, aged 6 months to 5 years attending the outpatient general clinics of Cairo University Children Hospital for non-infectious complaints. We excluded children with fever, signs of infection, history of antibiotic intake or hospitalization in the preceding month. The algorithm for laboratory testing is shown in .
Methods
Collection of Nasopharyngeal (NP) Secretion
We collected NP secretion by flocked swabs (Copan Flock Technologies, Brescia-Italy) on STGG medium (skim milk, tryptone, glucose, and glycerol); swab shaft was cut with scissors and transported to the laboratory within 30 minutes according to WHO guidelines.Citation5
Culture and AST
Each sample was vortexed for 10 seconds, and then 10 µL was streaked on a sheep blood agar plate (BAP). BAPs were incubated at 35°C in a CO2 enriched atmosphere using a candle jar for 18 to 24 hours; the rest of the sample was aliquoted and stored at −80ºC. Plates were examined and alpha-hemolytic colonies were tested for optochin susceptibility and bile-solubility.Citation6 Optochin-susceptible bile-soluble isolates were identified as S. pneumoniae and tested for antimicrobial susceptibility using Vitek-2™ (BioMérieux, Marcy-L’Etoile, France) and interpreted following standard guidelines.Citation7
Serotyping
DNA was extracted directly from specimens using the DNeasy blood and tissue kit (Qiagen, Valencia, CA) following the manufacturer’s instructions. A pneumococcus-specific real-time PCR targeting the lytA and Ply genes was performed as described previously.3 S. pneumoniae serotyping was performed on RT-PCR positive samples, using the Centers for Disease Control and Prevention Citation8 S. pneumoniae serotyping procedure, African set.Citation9 We conducted a sequential multiplex PCR on DNA extracted from biological samples. Forty-two primer pairs for serotypes 1, 2, 3, 4, 5, 6A/B/C, 7F/A, 7C/B, 8, 9V/A, 9N/F,10A,10F, 11A/D, 12A/F, 13, 14, 15A/F, 15B/C, 16F, 17F, 18A/B/C, 19A,19F, 20, 21, 22A/F, 23A, 23B, 23F, 24A/B/F, 25F/38, 31, 33A/F, 34, 35A/C, 35B, 35F, 35A, 38 and 39 were grouped into eight multiplex reactions. CpsA (pneumococcal capsular polysaccharide synthesis gene) primers were included in each reaction mix as a confirmatory test. The PCRs were performed in 25 µL volumes, with the mixture containing multiplex PCR Master Mix (Qiagen, Valencia, CA) and primers (0.2–0.5 mM each); 5 ul DNA extract was used in each PCR. Amplification was performed in a Seeamp Thermal Cycler (Seegene) under the following conditions: 95ºC for 15 min followed by 35 amplification cycles of 94ºC for 30 s, 54ºC for 90 s and 72ºC for 60 s. A final hold was performed at 72ºC for 10 min.
The PCR products were analyzed by gel electrophoresis on 2% agarose gels in TAE buffer. Gels were stained with ethidium bromide (0.5 mg/mL), and gel images were recorded. The sizes of the PCR products were determined by comparison with the molecular size standard as well as with the bands of the respective controls.
Forty-one clinical isolates of pneumococci representing different serotypes and serogroups were us ed as a control set. The serotypes included 1, 2, 3, 4, 5, 6A, 6C,7C,7F, 8, 9N, 9V, 10A, 10F,11A, 11B, 12F,13, 14, 15A, 15C, 16F, 17F, 18C, 19A, 19F, 20, 21, 22F, 23A, 23B, 23F, 24F, 31, 33F, 34, 35B, 35F, 35A, 38, 39.
S. pneumoniae from IPD
During the study period, we detected 40 S. pneumoniae from sterile body sites (27 from blood, 5 from CSF and 8 from pleural fluid) out of 7200 clinical specimens routinely submitted to our laboratory affiliated to Cairo University Hospitals which provide a tertiary care service to patients coming from different governorates all over Egypt. Of those 19 were detected by culture and their AST was done by VITEK-2. The other 21 specimens were culture-negative and detected only by PCR; this could be attributed to the antibiotic intake before the collection of microbiological cultures, especially before presenting to the hospital. For all 40 IPD samples, resistance to the penicillin and macrolides was studied by detection of the resistance genes PBP-2, mef and erm as follows.
Detection of Resistance Genes
Genotypic macrolide resistance (ermB and mef) and wild-type alleles of pbp2b typically associated with penicillin susceptibility were detected by CDC recommended multiplex real-time PCR protocol.Citation10 Briefly, primers’ sequences, concentrations and cycle conditions were selected according to Srinivasan et, al.Citation11 Universal Taqman Master mix (Thermo Fischer, Foster City, CA) was used in a 25ul total reaction volume and operated on a 96 CFX Biorad real-time PCR machine.
Data Analysis
Data were statistically described in terms of mean ± standard deviation (± SD), range, or frequencies (number of cases) and percentages when appropriate. All statistical calculations were done using computer programs SPSS (Statistical Package for the Social Science; SPSS Inc., Chicago, IL, USA) version 17 for Microsoft Windows.
Results
Detection of S. pneumoniae from NP Samples
We enrolled 334 children, aged 6 months to 5 years, among the attending children to the outpatient general clinics of Cairo University Children Hospital for non-infectious complaints. Male cases represented 118 (54.1%). By PCR, S. pneumoniae was detected in 217 (65%) most of them were isolated from male carriers who represent 54.1% of the children. Regarding the culture method, we isolated 110 S. pneumoniae out of 334 (32.9%).
Serotyping
The most frequently detected serotypes were serotypes 1, 6, 19F, 5, 18ABC, 14 and 19A (). The total vaccine coverage is 77.4% of typable S. pneumonia (), considering the PCV13 valency as follows: 1, 3, 4, 5, 6A, 6B, 7F, 9V,14, 18C, 19A, 19F, 23F.
Table 1 Number, % and PCV13 Coverage of the Identified NP Serotypes
Antimicrobial Susceptibility
The AST revealed low susceptibility level to all antimicrobials except for vancomycin, linezolid, levofloxacin and clindamycin (100%100%, 90.9% and 72.7%, respectively) ().
Table 2 Antimicrobial Susceptibility of S. pneumoniae Colonizing the Nasopharynx
Typing and AST of S. pneumoniae from IPD
Male patients represented 67.5% (27) out of 40 cases with IPD. Pediatric patients showed invasive pneumococcal disease more than the adult (92.5% Vs 7.5%). The commonest serotypes were 1, 18ABC, 6ABC and 5 (). The IPD isolates showed 100%, 100%, 94.7%, 89.5%, 79%, 84.2% and 78.9% susceptibility to vancomycin, linezolid, clindamycin, levofloxacin, penicillin, cefotaxim and erythromycin, respectively. In the DNA of 40 invasive S. pneumoniae, PBP2b (wild type), mef and erm genes were detected at 27.5%, 75% and 42.5%, respectively ( and ). () showed a comparison between antimicrobial susceptibility by vitek and the detected resistant genes by PCR for the 19 S. pneumoniae isolates from IPD. All S. pneumonia isolates which were sensitive to penicillin by MIC expressed the wild type of PBP-2b and were detected by PCR except two isolates. On the other hand, the resistant isolates to erythromycin were positive for erythromycin-resistant genes (mef2, erm) except two isolates that showed false-positive erythromycin resistance genes.
Table 3 Number, Percentage and PCV13 Coverage of Serotypes of 40 S. pneumoniae from IPD
Table 4 Antimicrobial Susceptibility of 19 S. pneumoniae Isolates from 19 IPD
Table 5 Detection of Resistance Genes in DNA of 40 S. pneumonia from IPD
Table 6 Comparison Between Antimicrobial Susceptibility by Vitek and the Detected Resistant Genes by PCR for the 19 Isolated S. pneumonia
Discussion
In this study, we identified the serotypes of S. pneumoniae NP carriage from healthy children or children with non-infectious diseases in two successive winter seasons. The most frequently detected serotypes from nasopharyngeal carriage were serotypes 1, 6, 19F, 5, 18ABC, 14 and 19A; and 77.4% of S. pneumoniae serotypes were covered by PCV-13.
The frequent carriage of ST 1, 6 and 19 F in the current study is in keeping with the recently published report from Egypt.Citation3 In contrast, ST 5, 14 and 18 were not detected in the older study, while ST 34 that was frequent in the previous study was not detected in the current study. The 77.4% vaccine coverage of NP S. pneumoniae in this study is in agreement with the reported 72.4% coverage in the previous study.Citation3
Surveillance of serotypes of S. pneumoniae is conducted in many countries. Our results are in broad agreement with studies on carriage in Egypt, Africa, the Middle East and other parts of the world.Citation12–Citation14
The temporal variation in serotypes occurs probably as a result of natural genetic transformation and bacterial interaction in the nasopharynx, which is rich in closely related microbial flora. This natural transformation has been known to play an important role in the evolution of the genomes of members of Genus Streptococcus.Citation4 As an example, Streptococcus mitis group that are common NP colonizers together with S. pneumoniae has long been recognized as a reservoir of antibiotic resistance genes for S. pneumoniae, as well as a source for capsule polysaccharide variation, leading to resistance and vaccine escape.Citation15
During the study period, we detected 19 S. pneumoniae isolates from IPD out of 7200 clinical specimens submitted to the laboratory. Additionally, we identified 21 S. pneumoniae by PCR from a sterile body site. The low number of IPD patients detected during the study period is consistent with previous results from Egypt.Citation16 In their hospital-based surveillance for laboratory-confirmed S. pneumoniae cases in children younger than 5 years, from 2008 to 2011, Draz et al isolated S. pneumoniae from 12 patients: four IPD and eight non-IPD out of more than 22,000 cultured specimen.Citation16 The low detection rate may be attributed to antibiotic intake before the collection of microbiological cultures, especially before presenting to the hospital. Antibiotics are available over the counter in Egyptian pharmacies, and self-medication and purchasing without medical prescriptions are common practice in Egypt.Citation17,Citation18
However, S. pneumoniae was more frequently identified from children with severe pneumonia in Egypt.Citation16,Citation19 In a study on 100 children with the severe community-acquired pneumonia admitted to ICU in Assiut (in southern Egypt), S. pneumoniae was isolated from 36% of the cases. Compared to other patients, pneumococcal pneumonia patients presented with more severe clinical picture (p<0.001) and resulted in higher mortality rate (P=0.45).Citation19 In contrast to our results, IPD is more frequently reported from other developing countries.Citation20
From pneumococcal IPD, the commonest serotypes were ST 1, 18ABC, 6 and 5; and the PCV13 coverage was 75% (). ST 1, 6, 18ABC and 5 were the commonest both in NP colonization and IPD, which proves the close relationship between colonization and IPD. NP colonization induces pro-inflammatory chemocytokines, along with damage to the respiratory epithelium caused by upper respiratory viral infection facilitates pneumococcal penetration of host tissues and progression to localized or invasive disease.Citation21 Multiple studies were in accordance with ours regarding the serogroups detected in IPD; ST1 had a significantly higher invasive disease potential in children aged 0–23 and 0–59 months,Citation22 serotypes 1 and 5 were likely to be about 3−4 times more invasive in children more than 2 years.Citation23 William et al declare that ST1 and 5 have commonly been viewed as “developing country” serogroups and cause 15–20% of IPD in Latin America, Asia, and Africa. In all these studies, the apparent differences in serotype distribution are influenced by age, population, geographic area, regional variations and time after PCV introduction.Citation24
Antimicrobial Susceptibility Testing (AST)
The nasopharyngeal isolates revealed low susceptibility to all antimicrobials except for vancomycin, linezolid, levofloxacin and clindamycin. Compared to results of Badawy et al,Citation3 the new series showed an increase in the percentage of susceptibility to penicillin from 27.4% to 32.7%, clindamycin from 64.5% to 72.7%, erythromycin from 50% to 55.5% and sulfamethoxazole/trimethoprim from 3.3% to 45.5%. This might be due to the infrequent utilization of these groups as previously reported.Citation25 In contrast, susceptibility to ceftriaxone markedly decreased from 99.8% to 54.5%, which is consistent with increased utilization of third-generation cephalosporins in Egypt,Citation25 and reported resistance to third-generation cephalosporins.Citation26 Our rates of resistance are higher than a report from Lithuania that showed 56.7% of S. pneumonia NP isolates were susceptible to all the antibiotics tested, and non-susceptibility to penicillin, erythromycin, clindamycin and trimethoprim/sulphamethoxazole was 15.8%, 21.3%, 16.9% and 27.3%, respectively.Citation27
However, invasive S. pneumoniae isolates showed lower rates of resistance compared to NP isolates. This is in accordance with a previous report that respiratory tract isolates showed more resistance to penicillin, TMP-SMX, cefuroxime and erythromycin, than invasive isolates, and were more likely to be multidrug resistant.Citation28 The Canadian Bacterial Surveillance Network also reported higher rates of penicillin NS for non-sterile compared with sterile isolates (16.6% versus 12.4%, P=0.005), with respiratory tract specimens constituting 60% of the non-sterile isolates.Citation29 Similarly, in Quebec, noninvasive isolates demonstrated higher rates of resistance to macrolides, penicillin and cephalosporins.Citation30 The « Tracking Resistance in the United States Today (TRUST) » program in the United States reported higher rates of resistance among respiratory isolates versus blood for penicillin, azithromycin, TMP-SMX and levofloxacin (P<0.0001 for all comparisons).Citation31
Nevertheless, resistance genes were frequently detected in our invasive S. pneumoniae isolates. Molecular detection of resistance mechanisms has been proved to correlate with AST and to be helpful in rapid detection of resistance of pathogens causing life-threatening infections.Citation32 Moreover, we could use this method in culture-negative IPD. Comparing conventional AST by VITEK-2 to the detection of resistance genes in 19 invasive S. pneumoniae, we found good agreement (78.9% and 89.5% for penicillin and erythromycin genes, respectively. However we encountered disagreement regarding penicillin resistance in 4 isolates; 2 susceptible isolates with negative wild type PBP-2b and 2 resistant isolates with positive wild-type PBP-2b. Penicillin resistant isolates giving positive results for the wild type PBP2b, might be explained by amino acid substitutions near the sequence encoding the downstream KTG motif which can confer PEN resistanceCitation11 or other mutations in different genes such as PBP2x and PBP1a.Citation33 On the other hand, penicillin-sensitive isolates giving negative result for the wild-type PBP2b need to be further studied and sequenced to uncover the unique genetic resistance profile of the Egyptian pneumococcal isolates that might have different substitutions at other sites. Regarding erythromycin, two susceptible isolates had false-positive mef-A and erm-B genes. This false positivity indicates the presence of similar resistance genes from other microorganisms in the clinical specimens, leading to cross-reactivity. Additionally, the mosaic genes that encode low-affinity PBP genes are products of recombination involving horizontal transfer from closely related species. Similarly, erm(B) and mef(A) macrolide-resistant genes in S. pneumoniae are highly homologous to genes in other Streptococcus species.Citation34 On the contrary, failure to detect the resistance genes (false negative) might be potentially conferred through other previously described mechanisms such as mutations in ribosomal genes encoding 23S rRNA and riboproteins L4 and L22.Citation35
Conclusion
The results of this study confirm the high carriage rate of S. pneumoniae among infants and children, and the substantial increase in emerging MDR strains in Egypt. As the PCV provides very good coverage to the S. pneumoniae serotypes causing colonization and IPD in Egypt, and considering the vulnerability of the younger age group and high-risk population, we recommend including pneumococcal conjugate vaccines in the national vaccination program in Egypt.
In response to the high toll of pneumococcal infections, the global community needs to recognize the impediments to vaccine introduction into Low Middle-Income Countries including Egypt. Improving PCV access could help decrease the incidence of pneumococcal infections and reduce the selection pressure for pneumococcal antimicrobial resistance.Citation2
Acknowledgments
The study of NP colonization was funded by an investigator initiated grant from Pfizer Medical to Professor Dr. Magda Badawy, Faculty of Medicine, Cairo University. Title: “Detection of the rate of nasopharyngeal carriage and serotypes of Streptococcus pneumoniae in Egyptian children less than five years”.
Disclosure
The authors report no conflicts of interest in this work.
References
- Ludwig E, Bonanni P, Rohde G, Sayiner A, Torres A. The remaining challenges of pneumococcal disease in adults. Eur Respir Rev. 2012;21(123):57–65. doi:doi:10.1183/09059180.0000891122379175
- Tricarico S, McNeil HC, Cleary DW, et al. Pneumococcal conjugate vaccine implementation in middle-income countries. Pneumonia. 2017;9(1). doi:doi:10.1186/s41479-017-0030-5
- Badawy M, El Kholy A, Sherif MM, et al. Serotypes of Streptococcus pneumoniae in Egyptian children: are they covered by pneumococcal conjugate vaccines? Eur J Clin Microbiol Infect Dis. 2017;36(12):2385–2389. doi:doi:10.1007/s10096-017-3071-z28744663
- Straume D, Stamsås GA, Håvarstein LS. Natural transformation and genome evolution in Streptococcus pneumoniae. Infect Genet Evol. 2015;33:371–380. doi:doi:10.1016/j.meegid.2014.10.02025445643
- Satzke C, Turner P, Virolainen-Julkunen A, et al. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine. 2013;32(1):165–179. doi:doi:10.1016/j.vaccine.2013.08.06224331112
- Croisier-Bertin D, Piroth L, Charles P-E, et al. Ceftaroline versus ceftriaxone in a highly penicillin-resistant pneumococcal pneumonia rabbit model using simulated human dosing. Antimicrob Agents Chemother. 2011;55(7):3557–3563. doi:doi:10.1128/AAC.01773-0921576444
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Second Informational Supplement Clinical and Laboratory Standards Institute; 2015.
- CDC, Ncird. Protocol for Multiplex PCR-S. pneumoniae SEROTYPING - clinical specimens and pneumococcal isolates - African set. 2010. doi:10.1128/JCM.02243-09
- Carvalho MDG, Pimenta FC, Jackson D, et al. Revisiting pneumococcal carriage by use of broth enrichment and PCR techniques for enhanced detection of carriage and serotypes. J Clin Microbiol. 2010;48(5):1611–1618. doi:doi:10.1128/JCM.02243-0920220175
- CDC, Ncird. Real-time PCR targets for Streptococcus species detection and antibiotic susceptibility in SPN*. doi:10.1371/journal.pone.0032169
- Srinivasan V, Du Plessis M, Beall BW, McGee L. Quadriplex real-time polymerase chain reaction (lytA, mef, erm, pbp2bwt) for pneumococcal detection and assessment of antibiotic susceptibility. Diagn Microbiol Infect Dis. 2011;71(4):453–456. doi:10.1016/j.diagmicrobio.2011.08.01721996097
- Wasfy MO, Pimentel G, Abdel-Maksoud M, et al. Antimicrobial susceptibility and serotype distribution of Streptococcus pneumoniae causing meningitis in Egypt, 1998-2003. J Antimicrob Chemother. 2005;55(6):958–964. doi:10.1093/jac/dki10115820983
- Shibl A, Memish Z, Pelton S. Epidemiology of invasive pneumococcal disease in the Arabian Peninsula and Egypt. Int J Antimicrob Agents. 2009;33(5):410.e1-410.e9. doi:10.1016/j.ijantimicag.2008.08.012
- Assefa A, Gelaw B, Shiferaw Y, Tigabu Z. Nasopharyngeal carriage and antimicrobial susceptibility pattern of streptococcus pneumoniae among pediatric outpatients at gondar university hospital, north west ethiopia. Pediatr Neonatol. 2013;54(5):315–321. doi:10.1016/j.pedneo.2013.03.01723680262
- Salvadori G, Junges R, Morrison DA, Petersen FC. Competence in streptococcus pneumoniae and close commensal relatives: mechanisms and implications. Front Cell Infect Microbiol. 2019. doi:10.3389/fcimb.2019.00094
- Draz IH, Halawa EF, Wahby G, Ismail DK, Meligy BS. Pneumococcal infection among hospitalized Egyptian children. J Egypt Public Health Assoc. 2015;90(2):52–57. doi:10.1097/01.EPX.0000465234.31794.b126154831
- Sabry NA, Farid SF, Dawoud DM. Antibiotic dispensing in Egyptian community pharmacies: an observational study. Res Soc Adm Pharm. 2014;10(1):168–184. doi:10.1016/j.sapharm.2013.03.004
- El-Hawy RM, Ashmawy MI, Kamal MM, et al. Studying the knowledge, attitude and practice of antibiotic misuse among Alexandria population. Eur J Hosp Pharm. 2017;24(6):349–354. doi:10.1136/ejhpharm-2016-00103231156970
- Aly SA, Elsherbiny NM, Ahmed SH, El-asheer OM. Prevalence of Streptococcus pneumoniae in Egyptian children. 2017;3(8):103–107.
- Manoharan A, Manchanda V, Balasubramanian S, et al. Invasive pneumococcal disease in children aged younger than 5 years in India: a surveillance study. Lancet Infect Dis. 2017;17(3):305–312. doi:10.1016/S1473-3099(16)30466-227956163
- Weiser JN, Ferreira DM, Paton JC. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol. 2018;16(6):355–367. doi:10.1038/s41579-018-0001-829599457
- Balsells E, Dagan R, Yildirim I, et al. The relative invasive disease potential of Streptococcus pneumoniae among children after PCV introduction: a systematic review and meta-analysis. J Infect. 2018;77(5):368–378. doi:10.1016/j.jinf.2018.06.00429964140
- Yildirim I, Hanage WP, Lipsitch M, et al. Serotype specific invasive capacity and persistent reduction in invasive pneumococcal disease. Vaccine. 2010;29(2):283–288. doi:10.1016/j.vaccine.2010.10.03221029807
- Hausdorff WP, Bryant J, Paradiso PR, Siber GR. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part I. Clin Infect Dis. 2000. doi:10.1086/313608
- Talaat M, Saied T, Kandeel A, et al. A point prevalence survey of antibiotic use in 18 hospitals in Egypt. Antibiotics. 2014;3(3):450–460. doi:10.3390/antibiotics303045027025755
- See I, Lessa FC, ElAta OA, et al. Incidence and pathogen distribution of healthcare-associated infections in pilot Hospitals in Egypt. Infect Control Hosp Epidemiol. 2013;34(12):1281–1288. doi:10.1086/67398524225613
- Stacevičiene I, Petraitiene S, Vaičiuniene D, Alasevičius T, Kirsliene J, Usonis V. Antibiotic resistance of Streptococcus pneumoniae, isolated from nasopharynx of preschool children with acute respiratory tract infection in Lithuania. BMC Infect Dis. 2016;16(1). doi:10.1186/s12879-016-1544-9
- Vanderkooi OG, McConnell A, Church DL, Kellner JD. Antimicrobial susceptibility of invasive and lower respiratory tract isolates of Streptococcus pneumoniae, 1998 to 2007. Can J Infect Dis Med Microbiol. 2009;20(4):139–144. doi:10.1155/2009/413749
- Powis J, McGeer A, Green K, et al. In vitro antimicrobial susceptibilities of Streptococcus pneumoniae clinical isolates obtained in Canada in 2002. Antimicrob Agents Chemother. 2004;48(9):3305–3311. doi:10.1128/AAC.48.9.3305-3311.200415328089
- Weiss K, et al. Genotypic characterization of macrolide-resistant strains of Streptococcus pneumoniae isolated in Quebec, Canada, and in vitro activity of ABT-773 and telithromycin. J Antimicrob Chemother. 2002;50(3):403–406. doi:10.1093/jac/dkf14612205066
- Karlowsky JA, Thornsberry C, Jones ME, Evangelista AT, Critchley IA, Sahm DF. Factors associated with relative rates of antimicrobial resistance among streptococcus pneumoniae in the United States: results from the TRUST Surveillance Program (1998–2002). Clin Infect Dis. 2003;36(8):963–970. doi:10.1086/37405212684907
- Nagai K. Evaluation of PCR primers to screen for Streptococcus pneumoniae isolates and beta-lactam resistance, and to detect common macrolide resistance determinants. J Antimicrob Chemother. 2001;48(6):915–918. doi:10.1093/jac/48.6.91511733479
- Diawara I, Nayme K, Katfy K, et al. Analysis of amino acid motif of penicillin-binding proteins 1a, 2b, and 2x in invasive Streptococcus pneumoniae nonsusceptible to penicillin isolated from pediatric patients in Casablanca, Morocco. BMC Res Notes. 2018;11(1). doi:10.1186/s13104-018-3719-5
- Fukushima KY, Yanagihara K, Hirakata Y, et al. Rapid identification of penicillin and macrolide resistance genes and simultaneous quantification of Streptococcus pneumoniae in purulent sputum samples by use of a novel real-time multiplex PCR assay. J Clin Microbiol. 2008;46(7):2384–2388. doi:10.1128/JCM.00051-0818463207
- Tait-Kamradt A, Davies T, Cronan M, Jacobs MR, Appelbaum PC, Sutcliffe J. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob Agents Chemother. 2000;44(8):2118–2125. doi:10.1128/AAC.44.8.2118-2125.200010898684