Abstract
Background
Worldwide, bacterial bloodstream infections (BSIs) constitute an important cause of morbidity and mortality in clinical settings. Due to the limited laboratory facilities in sub-Saharan Africa, poor diagnosis of BSIs results in poor clinical outcomes and leads to a risk of antimicrobial resistance. The present work was carried out to describe the microbiological features of BSIs using the data collected from Centre Hospitalier Universitaire de Kigali (CHUK).
Methods
A retrospective study was carried out at CHUK. The blood culture results of 2,910 cases – from adults, children and infants – were reviewed in the Microbiology service from October 2017 to October 2018. The following variables were considered: age, gender, admitting department, blood culture results, and antimicrobials sensitivity test results. Data were entered and analyzed using Microsoft Excel 2013.
Results
Twelve percent (341/2,910) of blood culture results reviewed were positive with 108 (31.7%) Gram positive bacteria and 233 (68.3%) Gram negative bacteria. The most prevalent pathogens were Klebsiella pneumoniae 108 (31.7%) and Staphylococcus aureus 100 (29.3%). This study revealed a high resistance to commonly prescribed antibiotics such as penicillin, trimethoprim sulfamethoxazole, and Ampicillin with 91.8, 83.3, and 81.8% of resistance, respectively. However, bacteria were sensitive to imipenem and vancomycin with 98.1 and 94.3% of sensitivity, respectively. The pediatrics and neonatology departments showed a high number of positive culture with 97/341 (28.4%), and 93/341 (27%) respectively. The overall prevalence of multidrug resistance was 77.1%.
Conclusion
The prevalence of bacterial pathogens in BSIs was found to be high. The antibiotic resistance to the commonly used antibiotics was high. Appropriate treatment of BSIs should be based on the current knowledge of bacterial resistance pattern. This study will help in formulating management of diagnostic guidelines and antibiotic policy.
Introduction
Bloodstream infections (BSIs) constitute one of the leading causes of mortality and morbidity worldwide.Citation1 Additionally, the infections with antibiotic-resistant bacteria have made the therapeutic options difficult due to the antibiotic misuse in humans, animals and agriculture.Citation2,Citation3 Although there is a paucity of data about BSIs in low- and middle-income countries (LMIC), it is believed that sub-Saharan Africa (SSA) could count a high prevalence of BSIs.Citation4–Citation6 However, BSIs remain undiagnosed due to limited diagnostic laboratory facilities resulting in poor clinical outcomes and the increasing risk of antimicrobial resistance.Citation7–Citation9 The rate of BSIs has been associated with hospitalization in intensive care units (ICU), insertion of foreign bodies, such as catheters, into blood vessels, lapses in hand washing, and non-adherence to infection prevention and control practices of health workers.Citation6,Citation10,Citation11
Worldwide, several population-based studies have reported a wide variation in incidence and prevalence of BSIs.Citation12,Citation13 A systematic review and meta-analysis of 22 studies about BSI during 1984–2006 in Africa, reported 58,296 patients receiving a blood culture request for BSI diagnosis of whom 2051 (13.5%) of 15,166 adults and 3527 (8.2%) of 43,130 children had BSI.Citation9 The most recent systematic review revealed a median prevalence of BSI at 14.6% (ranging from 3.4% to 38.2%) in Africa.Citation14 A specific characteristic of BSI in African countries is that many patients are co-infected with malaria, human immunodeficiency virus, or Mycobacterium tuberculosis.Citation15–Citation17
The most common microorganisms isolated in BSIs include on one hand bacteria such as Klebsiella pneumoniae, Escherichia coli, Enterobacter spp., Staphylococcus aureus, Coagulase Negative Staphylococcus (CoNS), Streptococcus pneumoniae, and Streptococcus pyogens and on other hand fungi such as Candida albicans and other Candida spp.Citation12,Citation18,Citation19 Among bacterial BSIs, antibiotic resistant strains are emerging with great speed, particularly among Gram-negative bacteria compromising effective treatment.Citation14,Citation20
In SSA, BSIs management is affected by the lack of bacteriological support for coordinated program that promotes the appropriate use of antibiotics and useful data for appropriate empirical antimicrobial treatment.Citation21,Citation22 In Rwanda, there are limited data of antimicrobial resistance in BSIs. Like several SSA countries, the lack of updated national guidelines for antibiotic use, absence of good laboratory facilities to perform blood culture and antimicrobial drug susceptibility tests lead to emergence and rapid spread of resistance.Citation23,Citation24
Despite the growing body of literature, there is wide geographic variation in causes of BSIs and in antimicrobial susceptibility patterns of identified microorganisms. Therefore, the present work was conducted to describe and extend the understanding of local laboratory characteristics of BSIs in Rwanda for better management of BSIs at Centre Hospitalier Universitaire de Kigali (CHUK) also known as Kigali University Teaching Hospital (KUTH). It is the largest hospital located in district of Nyarugenge in Kigali City. It is also the biggest referral hospital of the country with a capacity of 519 beds.
Materials and Methods
This was a retrospective study using available laboratory data from CHUK, Pathology Department from October 2017 to October 2018. Data were obtained from Microbiology unit log book of blood culture and antimicrobial sensitivity test results of both inpatients and outpatients. This study included laboratory records of all patients from whom BSIs were suspected. All laboratory data of blood culture samples with contamination and all laboratory records of bacterial isolates from blood culture samples that were not tested for antimicrobial susceptibility were excluded in the present study. Overall, s' the blood culture records of 2,910 patients were included and analyzed. The following variables were included: age, gender, admitting department, name of organism, antibiotics used for susceptibility testing and susceptibility results of each antibiotic tested. Microbial identification was performed following the laboratory standard operating procedures: the collected blood samples were incubated in BD BACTEC (Becton-Dickinson, Franklin Lakes, USA) bottles at 37°C and processed with the BACTEC system to identify bacteria. Positive samples for bacterial growth were sub-cultured on appropriate media after Gram stain results. Gram-positive cocci were cultured on Mannitol Salt Agar (MSA) and blood agar, and Gram-negative bacilli were cultured on MacConkey Agar and Xylose Lysine Deoxycholate Agar (XLD) media. In addition, identification of Gram-positive cocci species was done using catalase and coagulase tests. Gram-negative bacilli were identified by colony morphology. Additionally, biochemical tests were performed, including Triple Sugar Iron (TSI), Motility Indole Urea (MIU), and citrate tests to identify and differentiate Enterobacteriaceae species. The Antibiotic susceptibility testing was performed by the Kirby Bauer disk diffusion method, following laboratory protocol and the interpretation of the diameter of inhibition was done according to 2012 Clinical and Laboratory Standards Institute (CLSI) guidelines.
The ethical approval (Ref: EC/CHUK/7212/2018) to carry out the study was obtained from the Ethics Committee of Centre Hospitalier Universitaire de Kigali (CHUK). For retrospective studies, the informed consent to review the medical records of study participants was not required by the Ethics Committee. However, the data confidentiality and the right to privacy were respected in compliance with the Declaration of Helsinki.
Results
Out of 2,910 patient blood culture records reviewed, 1,530 (52.6%) were from males and 1,380 (47.4%) were from females. The majority of participants were in the age group 2–14 years, accounting for 29.4% (n=856) which was followed by 0–1-year age group (26.9%, n=785) and this trend was similar in both sexes.
From blood culture records, the results showed that 341/2,910 (11.71%) were positives. The number of pathogens isolated and represented by 10 types of bacteria was 341. Gram positive and Gram negative bacterial isolates constituted 108/341 (32%) and 233/341 (68%), respectively. Among Gram positive, S. aureus (29.3%) were predominant followed by Streptococcus spp. (2.1%) and Coagulase-negative Staphylococci (0.3%). The isolated Gram negative bacteria were K. pneumoniae (31.7%), followed by Acinetobacter spp. (14.3%), E. coli (13.2%), Salmonella typhi (5.6%), P. aeruginosa (2.9%), Citrobacter spp. (0.3%), and Proteus spp. (0.3%).
The distribution of blood culture samples based on the admitting department is summarized in the . Out of the 2,910 samples received, 1,012 (34.8%) were from the pediatrics department, 725 (25%) were from the medicine department and others. As shown in the , predominant culture isolates were from pediatrics department 97/341 (28.4%), followed by neonatology 93/341 (27%). K. pneumoniae was the most common isolate in the neonatology 46/108 (42.6%) and pediatrics 36/341 (10.6%). S. aureus and S. typhi were the most isolate in Internal medicine department with 28/100 (28%) and 11/19 (57.9%), respectively.
Figure 1 Service wise distribution of culture isolates.
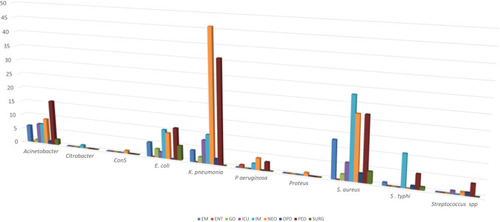
Table 1 Distribution of Blood Culture Results Based on Service
presents the susceptibility pattern of isolated pathogens. The antimicrobial susceptibility pattern of Enterobacteriaceae showed a high level of resistance of K. pneumoniae to the majority of antibiotics except imipenem and amikacin where it was sensitive at 100% and 88.1%, respectively. E. coli showed also a high resistance to the majority of antibiotics but was 100% sensitive to imipenem, clindamycin and piperacillin-tazobactam. S. typhi showed high level of sensitivity to amikacin (100%), ciprofloxacin (100%), clindamycin (100%), gentamycin (100%), imipenem (100%), ceftazidime (90%) and to amoxicillin-clavulanic acid (76.5%). However, the isolates were resistant to chloramphenicol (100%), trimethoprim–sulfamethoxazole (80%), piperacillin-tazobactam (66.7%), ampicillin (60%) and ceftriaxone (57.1%). Notably, all isolates were uniformly sensitive to imipenem (100%). In non-fermenting Gram negative rods, P. aeruginosa was sensitive to clindamycin (100%), imipenem (71.4%) and amikacin (66.7%). It showed 100% resistance to amoxicillin-clavulanic acid and penicillin, 75% piperacillin–tazobactam and gentamycin, 66.7% ceftazidime and piperacillin. Acinetobacter spp. showed high resistance to majority of antibiotics except gentamycin (100%), imipenem (100%), and amikacin (85.7%) where it was sensitive. Gram positive cocci showed a high level of resistance of S. aureus to amoxicillin-clavulanic acid (100%), penicillin (91.2%), trimethoprim – sulfamethoxazole (80%), and ampicillin (61.5%) but the isolates were sensitive to ceftriaxone (100%), vancomycin (96.3%), cefotaxime (88.9%), gentamycin (83.3%), and clindamycin (81.1%). CoNS showed also a high level of resistance to majority of antibiotics except vancomycin where it was 100% sensitive. Additionally, Streptococcus spp. showed a high level of resistance to majority of the antibiotics except ciprofloxacin (100%) and vancomycin (60%) where it was sensitive.
Table 2 Susceptibility Pattern of Isolated Pathogens at CHUK
The overall prevalence of multidrug resistance (MDR) (bacteria with non-susceptibility to at least one in three or more antimicrobial agents), was 77.1% (n=263). However, 6.5% (n=22) bacterial isolates were sensitive to all antibiotics tested. 194/233 (83.3%) Gram negative bacteria and 69/108 (63.9%) Gram-positive bacteria were multidrug resistant. 100% of CoNS and 62% (n=62) of S. aureus were multidrug resistant. Furthermore, 100% of Citrobacter, 100% of Proteus spp., 93.5% (n=101) of K. pneumoniae, 81.6% (n=40) of Acinetobacter spp. 73.3% (n=33) of E. coli, 63.2% (n=12) of S. typhi and 60% (n=6) of P. aeruginosa were multidrug resistant ().
Table 3 Multiple Antibiotics Resistance Pattern of Bacterial Isolates from Blood Culture
Discussion
It is well known that antibiotic- resistance represents a major public problem in SSA due to poor sanitation and weak public health system.Citation25 To our knowledge, this is the first study in Rwanda to assess the prevalence, and some factors associated with BSI and the prevalence of resistance in isolates by admission source using a large sample size.
The results of this present study are comparable with the observations made by Rani et al who reported 60.2% in males and 36.7% in females.Citation26 Similar studies have reported surprisingly the high prevalence of BSI in males without supporting explanations.Citation2 In fact, it has been reported that the high proportion of BSI originate from urinary tract infections, which are more common in females than in males.Citation27
This study revealed that the majority of blood culture results reviewed and the majority of culture isolates were from pediatrics department followed by neonatology. Consequently, a high proportion of participants were under 14-years-old and this trend was similar in both sexes. Children especially neonates are particularly vulnerable to BSIs mainly due to of their weak immune system; there is a clear evidence that immune dysregulation contributes to enhanced susceptibility of children to BSIs.Citation28 The maximum number of isolates (116) was in the age group of 0–1 year, accounting for 34% of the total 341 culture positive cases. In the age group 2–14 years, the number of isolates was 86 accounting for 25.2% culture positive cases. Similar findings have been reported by Nkrumah et al who reported culture positivity in infants up to 20.9%.Citation29
There is a very high variation of prevalence across the world. The isolate rate in this study was 11.7% (341/2910), which is similar with the reports in Zanzibar 14%Citation4 and Nigeria 13.1%.Citation30 However, the rate was high compared to the study conducted in NepalCitation31 and TanzaniaCitation32 with 7.2 and 7.7%, respectively but lower than the study in Ethiopia with 28%.Citation33
In the present study, the frequency of isolation of Gram negative bacteria (68.3%) was found to be higher than Gram positive bacteria (31.7%). Previous studies have reported similar trends with around 51% of Gram negative bacteria and 45% of Gram positive bacteria.Citation34 In contrast, some studies have shown a higher incidence of Gram positive bacteria than Gram negative bacteria.Citation35 This difference might be related to geographical variation and/or seasonal variation. It has been reported that temperatures are associated with substantially increased frequency of septicemia, particularly among clinically important Gram negative bacteria.Citation36 Most common bacteria isolated in this study were K. pneumoniae and S. aureus with 31.7% and 29.3%, respectively. Similar observations were reported in a number of studies.Citation15,Citation34
In this study, among the antibiotics used for susceptibility testing for Gram positive isolates, vancomycin showed highest (85.4%) activity. S. aureus was high sensitive to many antibiotics but was high resistant to commonly used antibiotics. This could be explained by the fact that Staphylococcal species could display completely different aggressiveness traits.Citation37 This study revealed that among the Gram negative isolates, imipenem showed the highest sensitivity (100%) which is consistent with the study conducted by Jhajhria et al and who also showed imipenem as most effective drug for Gram negative bacilli. Common antibiotics [erythromycin, penicillin, tetracycline, chloramphenicol and trimethoprim sulfamethoxazole] used for Gram negative showed the maximum resistance.Citation38
In East Africa, high levels of antimicrobial resistance to commonly used antibiotics have been reported including 50–100% of resistance to ampicillin, 20–47% of resistance to gentamicin and 46–69% of resistance to cephalosporins among Gram-negative infections.Citation39
Based on susceptibility tests in the present study, multi-drug resistance was observed in most of the isolates (77.1%). Similar study reported also a high multi drug resistance for blood culture isolates.Citation40
It is important to continually review and update the epidemiology of BSI mainly with respect to the antibiotic susceptibility pattern of the common pathogens, so that it is useful for prompt treatment of patients with BSIs.Citation41
Overall, this preliminary study is descriptive and informative: (i) it reveals a high resistance to the commonly prescribed antibiotics, (ii) shows the predominance of positive cultures in pediatrics department. However, some limitations have been noticed including the absence of correlating the antibiotic susceptibility pattern of isolated bacteria according to the age of patients, the lack of some data such as the distribution of inpatients versus outpatients and the lack of data on antibiotic consumption.
Conclusion
In this study, the prevalence of bacterial pathogens in blood stream infections was found to be high and was caused by both Gram positive and negative bacteria. The presence of drug resistance and multiple antimicrobial resistances bacteria to most commonly used antibiotics was high. Therefore, further studies are needed to draw consistent conclusions and recommendations to better support appropriate antimicrobial prescribing.
Data Sharing Statement
The datasets generated during this study including all relevant raw data, will be freely available to any scientist wishing to use them for non-commercial purposes, without breaching participant confidentiality.
Acknowledgments
We thank all staff of Microbiology laboratory at CHUK for both technical supports and helpful collaboration.
Disclosure
The authors report no conflicts of interest in this work.
References
- Hattori H, Maeda M, Nagatomo Y, et al. Epidemiology and risk factors for mortality in bloodstream infections: a single-center retrospective study in Japan. Am J Infect Control. 2018;46(12):75–79. doi:10.1016/j.ajic.2018.06.019
- Ntirenganya C, Manzi O, Muvunyi CM, Ogbuagu O. High prevalence of antimicrobial resistance among common bacterial isolates in a tertiary healthcare facility in Rwanda. Am J Trop Med Hyg. 2015;92(4):865–870. doi:10.4269/ajtmh.14-060725646259
- Muvunyi CM, Masaisa F, Bayingana C, et al. Decreased susceptibility to commonly used antimicrobial agents in bacterial pathogens isolated from urinary tract infections in Rwanda: need for new antimicrobial guidelines. Am J Trop Med Hyg. 2011;84(6):923–928. doi:10.4269/ajtmh.2011.11-005721633029
- Martin MJ, Thottathil SE, Newman TB. Antibiotics overuse in animal agriculture: a call to action for health care providers. Am J Public Health. 2015;105(12):2409–2410. doi:10.2105/AJPH.2015.30287026469675
- Leal HF, Azevedo J, Silva G, et al. Bloodstream infections caused by multidrug-resistant gram-negative bacteria: epidemiological, clinical and microbiological features. BMC Infect Dis. 2019;19(1):1–11. doi:10.1186/s12879-019-4265-z30606108
- Reddy EA, Shaw AV, Crump JA. Community-acquired bloodstream infections in Africa: a systematic review and meta-analysis. Lancet Infect Dis. 2010;10(6):417–432. doi:10.1016/S1473-3099(10)70072-4.Community-acquired20510282
- Onken A, Said AK, Melissa J, Jenum PA. Prevalence and antimicrobial resistance of microbes causing bloodstream infections in Unguja, Zanzibar. PLoS One. 2015;23:1–10. doi:10.1371/journal.pone.0145632
- Geldenhuys C, Chb MB, Sa F, et al. Central-line-associated bloodstream infections in a resource-limited South African neonatal intensive care unit. South Afr Med J. 2017;107(9):758–762. doi:10.7196/SAMJ.2017.v107i9.12124
- Archibald LK, Mcdonald LC, Nwanyanwu O, et al. A hospital-based prevalence survey of bloodstream infections in febrile patients in Malawi: implications for diagnosis and therapy. J Infect Dis. 1997;181:1414–1420. doi:10.1086/315367
- Nichols C, Maria L, Espinoza C, et al. Bloodstream infections and frequency of pretreatment associated with age and hospitalization status in Sub-Saharan Africa. Clin Infect Dis. 2015;61(Suppl 4):372–379. doi:10.1093/cid/civ730
- McLaws M-L. The relationship between hand hygiene and health care-associated infection: it ’ s complicated. Infect Drug Resist. 2015;8:7–18. doi:10.2147/IDR.S6270425678805
- Bassetti M, Righi E, Carnelutti A. Bloodstream infections in the intensive care unit. Virulence. 2016;7(3):267–279. doi:10.1080/21505594.2015.113407226760527
- Haque M, Sartelli M, Mckimm J, Bakar MA. Health care-associated infections – an overview. Infect Drug Resist. 2018;11:2321–2333. doi:10.2147/IDR.S17724730532565
- Laupland KB, Church DL. Population-based epidemiology and microbiology of community-onset bloodstream infections. Clin Microbiol Rev. 2014;27(4):647–664. doi:10.1128/CMR.00002-1425278570
- Buetti N, Atkinson A, Marschall J, Kronenberg A. Incidence of bloodstream infections: a nationwide surveillance of acute care hospitals in Switzerland 2008–2014. BMJ Open. 2017;7(3):1–4. doi:10.1136/bmjopen-2016-013665
- Marchello CS, Dale AP, Pisharody S, Rubach MP, Crump A. A systematic review and meta-analysis of the prevalence of community-onset bloodstream infections among hospitalized patients in Africa and Asia. Antimicrob Agents Chemother. 2020;64(1):1–16.
- Akoua-koffi C, Tia H, Plo JK, et al. Epidemiology of community-onset bloodstream infections in Bouaké, central Côte d’Ivoire. New Microbes New Infect. 2015;7(September2014):100–104. doi:10.1016/j.nmni.2015.06.00926442153
- Lochan H, Pillay V, Bamford C, Nuttall J, Eley B. Bloodstream infections at a tertiary level paediatric hospital in South Africa. BMC Infect Dis. 2017;17(1):1–9. doi:10.1186/s12879-017-2862-228049444
- Droz N, Hsia Y, Ellis S, Dramowski A, Sharland M, Basmaci R. Bacterial pathogens and resistance causing community acquired paediatric bloodstream infections in low- and middle- income countries: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2019;5:1–12.
- Duggan S, Leonhardt I, Kerstin Hunniger OK, Kurzai O. Host response to Candida albicans bloodstream infection and sepsis. Virulence. 2015;6(4):316–326. doi:10.4161/21505594.2014.98809625785541
- Van Schalkwyk E, Iyaloo S, Naicker SD, et al. Large outbreaks of fungal and bacterial bloodstream infections in a Neonatal Unit, South Africa, 2012–2016. Emerg Infect Dis. 2018;24(7):2012–2016. doi:10.3201/eid2407.171087
- Akova M. Epidemiology of antimicrobial resistance in bloodstream infections. Virulence. 2016;7(3):252–266. doi:10.1080/21505594.2016.115936626984779
- Opintan Japheth A, Newman MJ, Arhin RE, Donkor ES, Gyansa-lutterodt M, Mills-pappoe W. Laboratory-based nationwide surveillance of antimicrobial resistance in Ghana. Infect Drug Resist. 2015;8:379–389. doi:10.2147/IDR.S8872526604806
- Seboxa T, Amogne W, Abebe W, Tsegaye T. High mortality from blood stream infection in Addis Ababa, Ethiopia, is due to antimicrobial resistance. PLoS One. 2015;12:1–14. doi:10.1371/journal.pone.0144944
- Kariuki S, Dougan G. Antibacterial resistance in sub-Saharan Africa: an underestimated emergency. Ann N Y Acad Sci. 2014;1323(1):43–55. doi:10.1111/nyas.1238024628272
- Rani R, Chaitanya S, Rajappa S, et al. Retrospective analysis of blood stream infections and antibiotic susceptibility pattern of gram negative bacteria in a Tertiary Care Cancer Hospital. Int J Med Res Health Sci. 2017;6(12):19–26.
- Al-hasan MN, Lahr BD, Eckel-passow JE, Baddour LM. Epidemiology and outcome of Klebsiella Species bloodstream infection: a Population-Based Study. Mayo Clin Proc. 2010;85(2):139–144. doi:10.4065/mcp.2009.041020118389
- Bard JD, Tekippe E, Kraft CS. Diagnosis of bloodstream infections in children. J Clin Microbiol. 2016;54(6):1418–1424. doi:10.1128/JCM.02919-1526818669
- Nkrumah NO, Labi AK, Addison NO, Ewuramma J, Labi M, Mensah GA. Trends in paediatric and adult bloodstream infections at a Ghanaian referral hospital: a retrospective study. Ann Clin Microbiol Antimicrob. 2016;15:1–10. doi:10.1186/s12941-016-0163-z26786830
- Nwadioha I, Nwokedi E, Odimayo MS. A review of bacterial isolates in blood cultures of children with suspected septicemia in a Nigerian tertiary Hospital. Afr J Microbiol Res. 2010;4(4):222–225.
- Prakash Simkhada SR, Lamichhane S, Subedi S, Shrestha UT. Bacteriological profile and antibiotic susceptibility pattern of blood culture isolates from patients visiting Tertiary Care Hospital in Kathmandu, Nepal. Glob J Med Res Microbiol Pathol. 2016;16:1.
- Mahende C, Ngasala B, Lusingu J, et al. Bloodstream bacterial infection among outpatient children with acute febrile illness in north ‑ eastern Tanzania. BMC Res Notes. 2015;8:1–8. doi:10.1186/s13104-015-1178-925645429
- Wasihun AG, Wlekidan LN, Gebremariam SA. Bacteriological profile and antimicrobial susceptibility patterns of blood culture isolates among febrile patients in Mekelle Hospital, Northern Ethiopia. Springerplus. 2015;4. doi:10.1186/s40064-015-1056-x
- Negussie A, Mulugeta G, Bedru A, et al. Bacteriological profile and antimicrobial susceptibility pattern of blood culture isolates among septicemia suspected children in Selected Hospitals Addis Ababa, Ethiopia. Int J Biol Med Res. 2016;6(1):4709–4717.
- Terfa KK, Dufera TB, Tinsae KMH, et al. Assessment of bacterial profile and antimicrobial resistance pattern of bacterial isolates from blood culture in Addis Ababa Regional Laboratory, clinical microbiology: open access. Clin Microbiol. 2018;7(2):1–6. doi:10.4172/2327-5073.1000312
- Eber MR, Shardell M, Schweizer ML, Laxminarayan R, Perencevich EN, Spellberg B. Seasonal and temperature-associated increases in gram-negative bacterial bloodstream infections among hospitalized patients. PLoS One. 2011;6(9):5–10. doi:10.1371/journal.pone.0025298
- Săndulescu O, Bleotu C, Matei L, et al. Comparative evaluation of aggressiveness traits in staphylococcal strains from severe infections versus nasopharyngeal carriage. Microb Pathog. 2016;102:45–53. doi:10.1016/j.micpath.2016.11.00627856272
- Jhajhria A, Yadav AK, Parihar G, Gupta PS. Bacteriological profile and antimicrobial susceptibility of blood culture in a tertiary care hospital Ajmer. Int J Med Health Res. 2018;4(6):7–11.
- Lucas A, Abraham M, Patrick O, Juliet M-A, Bebell Lisa BY. A review of antimicrobial resistance in East Africa. Afr J Lab Med. 2016;5(1):1–6.
- Zenebe T, Kannan S, Yilma D, Beyene G. Invasive bacterial pathogens and their antibiotic susceptibility patterns in jimma specialized hospital, Southwest Ethiopia. Ethiop J Health Sci. 2011;21(1):1–8. doi:10.4314/ejhs.v21i1.69038
- Coulter S, Roberts JA, Hajkowicz K, Halton K. The use of bloodstream infection mortality to measure the impact of antimicrobial stewardship interventions. Infect Dis Rep. 2017;9:8–12. doi:10.4081/idr.2017
