Abstract
Background
Recent reports of the rapid evolution of bacterial resistance in India require urgent antibiotic stewardship programs. This study aimed to define the magnitude and pattern of resistance of bacterial pathogens to guide empirical therapy.
Methods
We prospectively collected consecutive, clinically significant, and nonduplicate bacterial isolates from each patient from two hospitals in Ujjain, India. The antibiotic susceptibility of the bacteria was tested using a disc diffusion method as recommended by the Clinical and Laboratory Standards Institute.
Results
A total of 716 pathogens were isolated from 2568 patients (median age, 25 years; range, 0 days to 92 years). Gram-negative infections were predominant (62%). The isolated pathogens included Staphylococcus aureus (n = 221; 31%), Escherichia coli (n = 149; 21%), Pseudomonas aeruginosa (n = 127; 18%), and Klebsiella pneumoniae (n = 107; 15%). Common diagnoses included abscesses (56%), urinary tract infections (14%), blood stream infections (10%), pneumonia (10%), and vaginal infections (10%). In E. coli isolates, 69% (95% confidence interval [CI] 61.6–76.6) were extended-spectrum β-lactamase (ESBL) producers and 41% (95% CI 31.6–50.5) of K. pneumoniae isolates were ESBL producers. These isolates had a high resistance to fluoroquinolones and β-lactams, except for imipenem and piperacillin-tazobactam. Salmonella typhi remained sensitive to third-generation cephalosporins. Methicillin-resistant S. aureus (MRSA) constituted 30% of all S. aureus isolates and showed resistance to ciprofloxacin (81%), cotrimoxazole (76%), and levofloxacin (60%).
Conclusion
Our results showed a high prevalence of ESBL among Gram-negative bacterial isolates and a high prevalence of MRSA among S. aureus isolates. Carbapenems provided the broadest coverage for Gram-negative bacteria, while glycopeptides were the most effective against MRSA; however, both classes of drugs need to be used judiciously. This study will help in planning future antibiotic stewardship programs.
Introduction
Antibiotic resistance is a global public health problem.Citation1,Citation2 The foundation of modern medicine is built on the availability of effective antibiotics, especially in economically deprived areas of the world where the disease burden due to bacterial infections remains high. Antibiotic resistance is predominantly fueled by antibiotic use.Citation3 The regular introduction of new antibiotic classes over the years has partly masked the problem of increasing resistance. However, this is no longer the case today because the pipeline for newer antibiotics is nearly empty.Citation4 Therefore, we need to preserve the currently available antibiotics for use by future generations.Citation1,Citation2 The World Health Organization and European Commission have recognized the importance of studying the emergence and determination of resistance and the need for control strategies. The need for strategies to control antibiotic resistance is greater in resource constraint settings because antibiotic resistance puts further strain on an already fragmented health care system in low and middle-income countries.Citation5
One recent eye opener is the spread of Enterobacteriaceae, with resistance to carbapenem conferred by New Delhi metallo-β-lactamase 1 (NDM-1).Citation6 NDM-1 received extensive media coverage for two reasons, ie, the bacteria carrying the NDM-1 gene are resistant to all antibiotics except tigecycline and colistin, and were rapidly transmitted across national borders.Citation6
In India, rapid evolution of bacterial resistance may be due to a complex interaction of several factors such as higher burden of infectious disease, treatment uncertainty, lack of treatment guidelines, inadequate access to standard laboratory facilities, self-medication, prescription based on availability, government support to pharmaceutical industries, market forces, antibiotics prescribed by unqualified health professionals, less strict law enforcement, fragmented public health system, poor population-wide insurance coverage, inadequate adherence to universal hygiene and infection control measures, and to low population-wide education level.Citation7–Citation12 Antibiotic stewardship programs are thus urgently needed in India.Citation8,Citation10
Antibiotic susceptibility surveillance is fundamental for creating an antibiotic stewardship program. Thus, we set up a surveillance system in two hospitals in Ujjain, India, with the aim of defining resistance magnitude and patterns of bacterial pathogens and providing locally applicable data to guide empirical therapy.
Materials and methods
This prospective study was conducted over a period of 15 months from November 2007 to February 2009 in Ujjain, India.
Study settings
The study sites were two hospitals, ie, a 570-bed teaching hospital attached to RD Gardi Medical College and a 350-bed nonteaching hospital. Both hospitals cater predominantly for rural populations from the villages surrounding Ujjain city. In both hospitals, most admissions (89%–91%) to the medical and intensive care units are made on an emergency basis, whereas admissions into surgical units are made on an elective or emergency basis.
Collection of samples and study participants
We prospectively collected consecutive, nonduplicate, single patient samples. Only “clinically significant” samples (from patients with presumed infections) were sent for culture. The following infections and corresponding samples were included in the study: abscesses (pus/secretions and swabs from skin and soft tissue infections), post-surgery or traumatic wounds, and burns, and ear discharge (in clinically proven serous otitis media), urinary tract infections (mid-stream clean catch urine or urine from a catheter), blood stream infections (in cases of clinical sepsis), pneumonia (induced sputum and/or bronchoalveolar lavage), and vaginal infections (high vaginal swab). Isolates from the screening procedures and samples for fungal, mycobacterial, and anaerobic bacterial cultures were not included in the study.
The admitting consultants were requested to send clinically relevant samples for culture from all patients suspected of having a bacterial infection. The following demographic information was collected for all the patients: age, gender, family size, education level (of adult patients or of a child patient’s mother), breadwinner’s occupation, reported history of antibiotics received in the past two weeks, and reported hospitalizations in the past two weeks.
Participating departments in both hospitals included pediatrics, general medicine, general surgery, obstetrics and gynecology, ear, nose and throat, orthopedics, chest medicine, adult medicine intensive care unit, and neonatal intensive care unit.
Identification, antibiotic susceptibility testing, and definitions of resistance
Within four hours of receipt at the laboratory, all the samples were plated on blood agar and MacConkey agar medium (HiMedia Laboratories Pvt, Ltd, Mumbai, India). Pathogenic bacteria were identified using standard conventional microbiological methods.Citation13 Antibiotic sensitivity testing was performed using the Kirby-Bauer disc diffusion method on Mueller-Hinton agar plates. The disc strengths were as recommended by the Clinical and Laboratory Standards Institute (CLSI) at the time of the study. CLSI interpretive criteria for susceptibility and resistance were followed.Citation14 Antibiotic sensitivity testing quality control was performed using the following reference strains: Escherichia coli ATCC (American Type Culture Collection) 25922, Klebsiella pneumoniae ATCC 70063, Pseudomonas aeruginosa ATCC 27853, Enterococcus faecalis ATCC 29212, and Staphylococcus aureus ATCC 29213. Intermediate sensitive isolates of Gram-negative bacteria were counted as resistant in the calculations.
Extended-spectrum β-lactamase (ESBL) production was detected using a double-disc synergy test.Citation15 The presence of ESBL was assayed using the following antibiotic discs: cefotaxime 30 μg, cefotaxime/clavulanic acid 30/10 μg, ceftazidime 30 μg, and ceftazidime/clavulanic acid 30/10 μg (HiMedia Laboratories Pvt, Ltd, Mumbai, India). According to the CLSI criteria for ESBL detection, each isolate with an inhibition zone diameter of ≤22 mm for ceftazidime or ≤27 mm for cefotaxime was considered to be a potential ESBL producer or screen positive. A zone diameter increase of ≥5 mm for either antimicrobial agent when tested in combination with clavulanic acid versus when tested alone was considered as an ESBL-producing organism. K. pneumoniae ATCC700603 (positive control) and E. coli ATCC25922 (negative control) were used for quality control in the ESBL tests.Citation14
For S. aureus isolates, screening for methicillin resistance was performed using a cefoxitin disc screen test and 6 g/mL oxacillin in Mueller-Hinton agar supplemented with NaCl (4% w/v; 0.68 mol/L) according to the CLSI guidelines.Citation14 Multidrug-resistant isolates were defined as isolates having coresistance to at least three antibiotic groups.Citation16 The ethics committee of RD Gardi Medical College approved the study (approval number 41/2007). The cultures were performed without any cost to the patients, and the results were made available to each patient’s physician.
Statistical analysis
The data were entered in EpiData Entry (version 3.1) software and then transferred to Stata 10.0 for further analysis (Stata Corporation, College Station, TX). Descriptive statistics were used.
Results
Patient demographics and distribution within departments or wards
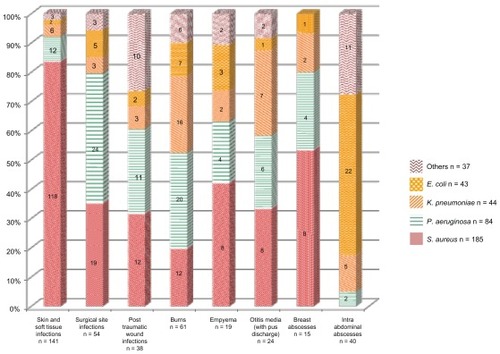
The main characteristics of patients enrolled in the study are shown in . A total of 716 pathogens were isolated from 2568 patients. Thus, the culture positivity rate was 28% (95% confidence interval [CI] 26.1–29.6). The median patient age was 25 (range, from 0 days to 92 years), and the median age of men and women was 23 and 26 years, respectively. Most samples (95%) were sent from wards while the remaining samples were sent from intensive care units. The most common diagnoses () were abscesses (56%), urinary tract infections (14%), blood stream infections (10%), pneumonia (10%), and vaginal infections (10%).
Table 1 Demographic characteristics of the 901 pediatric and 1667 adult patients from whom 716 pathogenic bacteria were isolated in Ujjain, India
Table 2 Distribution of commonest four pathogens per site of infection in surveillance study in two hospitals, Ujjain, India
Most common pathogens
The distribution of the four most common pathogens by infection site is shown in . Details of site of infection and proportion of different bacteria (n = 393) isolated from abscesses are shown in . Gram-negative infections were predominant (62%). However, the most commonly isolated pathogen was S. aureus (n = 221; 31%), followed by E. coli (n = 149; 21%), P. aeruginosa (n = 127; 18%), and K. pneumoniae (n = 107; 15%). S. typhi was isolated most frequently from blood stream infections (21%). E. coli constituted 66% of the urinary tract infection isolates. In pneumonia, P. aeruginosa was the most commonly isolated organism (n = 22; 30%), followed by K. pneumoniae (n = 21; 28%) and S. aureus (n = 10; 14%). Gram-negative organisms were isolated from the bronchoalveolar lavage samples from adults or from the pneumonia with sepsis samples from neonates. Gram-negative E. coli, K. pneumoniae, and P. aeruginosa constituted 75% of the organisms isolated from vaginal infection.
Antibiotic susceptibility testing of common Gram-negative bacteria
A total of 447 Gram-negative pathogens were isolated. The in vitro antibiotic susceptibility of the common Gram-negative bacteria is shown in . Remarkably high resistance for β-lactam antibiotics (range 72%–97%) and fluoroquinolones (range 51%–95%) was observed for the three most common Gram-negative bacteria, ie, E. coli, P. aeruginosa, and Klebsiella. E. coli showed the highest susceptibility (98%) to imipenem followed by piperacillin-tazobactam (85%). The same antibiotics were also active against other Enterobacteriaceae, Klebsiella, Pseudomonas, and Proteus species. S. typhi showed good sensitivity to the third-generation cephalosporins, ie, ceftriaxone (86%) and ceftazidime (88%), as compared with fluoroquinolones, ie, ciprofloxacin (31%) and ofloxacin (28%, ).
Table 3 Spectrum of activity of 20 antimicrobials against five most prevalent causes of Gram-negative infections in a surveillance study in two hospitals, Ujjain, India
The ESBL rates were 69% (95% CI 61.6–76.6) for E. coli and 41% (95% CI 31.6–50.5) for K. pneumoniae. Among E. coli, P. aeruginosa, and Klebsiella, coresistance to cephalosporins and fluoroquinolones was 51.2% (95% CI 44.8–58.3). The multidrug-resistant rate among Gram-negative bacteria was 17.6% (95% CI 14.1–21.2). The multidrug-resistant rates were highest for E. coli (21%; 95% CI 14.8–28.1), followed by K. pneumoniae (19%; 95% CI 11.1–26.1) and P. aeruginosa (17%; 95% CI 9.9–23.0). There were three pan-resistant isolates (for the tested antimicrobial drugs), one each for E. coli, P. aeruginosa, and K. pneumoniae.
Antibiotic susceptibility testing of common Gram-positive bacteria
The in vitro antibiotic susceptibility pattern of 154 methicillin-sensitive S. aureus (MSSA) isolates is shown in . Resistance to commonly used oral antibiotics, including ampicillin (86%), amoxicillin-clavulanate (50%), cotrimoxazole (38%), ciprofloxacin (49%), and erythromycin (9%) was noted in MSSA isolates. Because physicians commonly coprescribe amikacin in our study setting to treat serious S. aureus infections, we studied coresistance of S. aureus to a combination of amikacin with different classes of antibiotics. Coresistance for amikacin was observed for ampicillin (6%), amoxicillin-clavulanate (5%), ciprofloxacin (4%), ceftriaxone (1%), and chloramphenicol (1%). Coresistance to ciprofloxacin and erythromycin was 7%.
Table 4 Spectrum of activity of 25 antimicrobials against three most prevalent causes of Gram-positive infections in surveillance study in two hospitals, Ujjain, India
MSSA isolates did not show resistance to vancomycin, teicoplanin, linezolid, or clarithomycin. However, high susceptibility was also noted for cefoxitin (93%), amikacin (94%), clindamycin (92%), gentamicin (90%), and chloramphenicol (86%).
MRSA constituted 30% of all S. aureus isolates. The antibiotic susceptibility pattern of the 67 MRSA isolates is shown in . The MRSA isolates showed resistance to ciprofloxacin (81%), cotrimoxazole (76%), levofloxacin (60%), erythromycin (28%), and doxycycline (27%). Moreover, MRSA showed coresistance to levofloxacin and amikacin (27%), doxycycline and levofloxacin (27%), ciprofloxacin and amikacin (24%), ciprofloxacin and erythromycin (22%), doxycycline and amikacin (16%), and chloramphenicol with amikacin (9%). No resistance was noted for clindamycin and amikacin.
Coagulase-negative staphylococci (n = 20) were regarded as pathogens after considering the clinical condition of a patient and when cultured in paired blood samples. Methicillin resistance was 89%. A high proportion of resistance was observed for chloramphenicol (88%), cotrimoxazole (82), tetracycline (76%), doxycycline (68%), and clindamycin (54%).
Discussion
To the best of our knowledge, this is the first surveillance study that examined the antimicrobial susceptibilities of common pathogens in a rural resource-poor setting in central India. In India, most populations reside in and seek health care in similar settings. However, previously reported studies have been performed in larger metropolitan cities.Citation11,Citation12 The present study showed that the common pathogens were S. aureus (31%), E. coli (21%), P. aeruginosa (18%), and K. pneumoniae (15%). The ESBL rate of E. coli was 69% (95% CI 61.6–76.6), while that of K. pneumoniae was 41% (95% CI 31.6–50.5). MRSA constituted 30% of all S. aureus isolates. To improve the clinical outcome of patients in resource-poor settings, treatment guidelines for empirical therapy need to be formulated. These guidelines should ideally be aligned with local susceptibility patterns. Availability of this information will help clinicians select appropriate and effective therapy and reduce the incidence of drug-resistant bacteria.
In this study, abscesses, pneumonia, and urinary tract infection were responsible for 80% of all the culture-positive infections. E. coli, P. aeruginosa, K. pneumoniae, and S. aureus were responsible for 85% of culture-positive infections. Similar patterns of bacterial isolates are noted from other resource-constrained countries,Citation17,Citation18 including India.Citation11,Citation12 The SENTRY Antimicrobial Surveillance Program in the Asia-Pacific also reported S. aureus as the most common organism, followed by P. aeruginosa and E. coli.Citation19,Citation20
P. aeruginosa was the most common organism isolated from pneumonia and the second most common organism isolated from abscesses. It remains the leading cause of health care-associated infections worldwide, especially in patients admitted to intensive care units. The organism is ranked second only to S. aureus as the most common cause of health care-associated infections in most European intensive care units.Citation21 A single tertiary care center reported a three-fold increase in the number of P. aeruginosa isolates in India during the study period of 2007–2008 along with decreasing sensitivity to meropenem (from 64% in 2007 to 35% in 2008).Citation11 Decreasing sensitivity to third-generation cephalosporins and fluoroquinolones is also well established in India.Citation11,Citation12 Most of the isolates of P. aeruginosa in the present study could be from health care-associated infections. The susceptibility rates found in the present study (13% for ceftazidime, 37% for ciprofloxacin) are similar to those observed in other studies from India.Citation22,Citation23 Varghese et alCitation12 reported 70% resistance to fluoroquinolones and other second-line antipseudomonal drugs such as piperacillin/tazobactam (42%), cefoperazone/sulbactam (40%), and cefpirome (54%).
The third generation cephalosporins effective against Pseudomonas aeruginosa are ceftazidime and cefaperazone. Nearly 87% of the isolates in this study are resistant to Ceftazidime. This may be a local phenomenon.
Carbapenems, colistin, polymyxin B, or a combination of ceftazidime and an aminoglycoside can be used for the empirical treatment of Pseudomonas infections.Citation11,Citation24 Increasing carbapenem resistance is a global challenge,Citation20 but, fortunately, the phenomenon is rare in our setting.
Widespread resistance of S. typhi to commonly used oral antibiotics is a serious clinical and a public health challenge. In the present study, S. typhi showed high resistance to ampicillin, cotrimoxazole, and ciprofloxacin. This phenomenon has been reported in other studies, discussed in a recent review,Citation25 which suggested that fluoroquinolones and third-generation cephalosporins are equally effective treatment options for Salmonella infections.Citation25 In the present study, however, the sensitivity to third-generation cephalosporins remained high (86%–88%), indicating that they are a better treatment option than fluoroquinolones. However, minimum inhibitory concentrations for fluroquinolones and third-generation cephlosporins would better guide therapy.
It is widely accepted that the first-line empirical treatment for uncomplicated urinary tract infection should be cotrimoxazole if resistance rates of urinary E. coli to cotrimoxazole are <20%. If cotrimoxazole resistance is >20%, fluoroquinolone, nitrofurantoin, or fosfomycin are recommended.Citation26 In the present study, resistance to cotrimoxazole was documented to be >80%, which is in accordance with the high-resistance rates recorded in other Indian studies.Citation27 Most patients at the two study sites were treated with norfloxacin in outpatient clinics.Citation28 Because quinolones select more antibiotic-resistant strains and nitrofurantoin shows good activity against E. coli, nitrofurantoin would serve as better alternative first-line drug for the treatment of uncomplicated urinary tract infection in this setting. However, nitrofurantoin is not aggressively marketed and has supply-related problems. Treatment options for complicated urinary tract infection need to be individualized based on culture reports.
ESBL production was 69% in E. coli isolates and 41% in K. pneumoniae isolates. The above pattern of resistance is a clue toward other possible mechanisms of resistance, including AmpC production. Studies performed in India have shown that plasmid-borne and chromosomally mediated AmpC-and cephalosporinase-producing pathogens are common in resistant E. coli and K. pneumoniae isolates.Citation22,Citation29 The reported prevalence of ESBL-producing Gram-negative isolates in various hospitals in India is in the range of 19%–60%.Citation22
Prevalence of ESBL-producing Gram-negative isolates in various hospitals in India is in the range of 19%–60%. Cabapenem resistance is reported to be 5.3%–59% in various metropolitan tertiary care hospitalsCitation22
A study performed in a tertiary care hospital in Hyderabad by Subbalaxmi et alCitation11 showed that only 8% E. coli isolates were sensitive to ceftriaxone, a frequently used empirical antibiotic. Sensitivity to a combination of β-lactam β-lactamase inhibitors like cefaperazone/sulbactam and piperacillin/tazobactam was 59% and 61%, respectively. This study showed a high prevalence of ESBL in an urban set-up.Citation11 A high prescribing rate of fluroquinolones was documented in an outpatient study on antibiotic prescribing linked with diagnosis in the same settings.Citation28 The rate of fluoroquinolone use determines the resistance rates for both fluoroquinolones and third-generation cephalosporins.Citation30 This finding might explain the high rate of coresistance (51.2%) to third-generation cephalosporins and ciprofloxacin documented in the present study.
Among the Gram-positive organisms, S. aureus and coagulase-negative staphylococci were the most common organisms. MRSA constituted 30% of all S. aureus isolates. The antibiotic susceptibility pattern of MRSA showed resistance to commonly used antibiotics, which has been documented in Southeast AsiaCitation17,Citation18 and India.Citation11,Citation12 Varghese et alCitation12 in an Indian study performed at a tertiary care center in Manipal reported a MRSA rate of 35%. Increased resistance of MRSA isolates to almost all antibiotics over a one-year period was observed in this study.Citation12 They reported a high proportion of resistance of MRSA isolates to the commonly used first-line antibiotics, such as cotrimoxazole (96%), ciprofloxacin (85%), erythromycin (83%), and tetracyclines (72%). Our results showed better susceptibility to the above antibiotics except ciprofloxacin. These differences in the results could be due to the differences in geographical areas and settings. The resistance rates for ciprofloxacin in the two studies are quite similar (81% in our study versus 85% in the referenced study).Citation12 This finding could be because of high outpatient use of quinolones in our settings.Citation28 Other hospitals (predominantly urban) in India have reported MRSA rates of 8%–71%.Citation22
The main strength of this survey is that it provides much needed data for evidence-based discussion on prudent antibiotic prescribing in India. The data have links to clinical infections, information that is usually minimal or absent in laboratory-based studies. Our study has limitations. Because of the limited sample size, we were unable to detect uncommon forms of resistance, such as carbapenem resistance and vancomycin-resistant enterococci. The “gold standard” for antibiotic susceptibility reporting is minimum inhibitory concentration testing; this was not performed because we wanted to perform the tests that are normally available in this type of setting. Hospitalization durations were not recorded; thus, it was not possible to distinguish between health care-associated infections and community-acquired infections. The likelihood that the isolated organism is a colonizing bacterium should be considered in such studies. However, in the present study, because the inclusion criterion was a patient with a suspected infection, this possibility was minimized.
Conclusion
In conclusion, this is the first surveillance study of antimicrobial susceptibility performed in a rural resource-poor setting in central India. The results define the occurrence of pathogens and suggest the leading antimicrobial resistance mechanisms. Carbapenems provide the broadest coverage for Gram-negative bacteria and glycopeptides are most effective against MRSA; however, both these classes of drugs need to be used judiciously. Antimicrobial resistance surveillance is necessary for both day-to-day clinical decisions and to reflect local, regional, and national trends. Comprehensive surveillance programs linked to antimicrobial prescription need to be prioritized to set up effective infection control measures. The data from this study will be useful for planning antibiotic stewardship programs in India.
Acknowledgments
This study is funded by the Swedish Research Council and the Asia Link. We thank the Erasmus Mundus External Cooperation Window lot 15 and Swedish Research School for Global Health for scholarships to Ashish Pathak for his doctoral studies. The authors also wish to thank Rama Iyer and Binita Singh for contribution to microbiological analysis. We thank all the patients and staff for their cooperation during data collection. We also thank JK Sharma, Dean, and VK Mahadik, Medical Director, RD Gardi Medical College, for administrative support during the project.
Disclosure
The authors declare no conflict of interest in relation to this paper.
References
- World Health OrganisationImproving the containment of antimicrobial resistanceGeneva, SwitzerlandWorld Health Organisation2005 Available from: http://www.searo.who.int/LinkFiles/BCT_Regional_Strategy_ARM_ver31032010.pdfAccessed March 5, 2012
- CarsOHogbergLDMurrayMMeeting the challenge of antibiotic resistanceBr Med J2008337a143818801866
- GoossensHFerechMVander SticheleRElseviersMOutpatient antibiotic use in Europe and association with resistance: a cross-national database studyLancet2005365945957958715708101
- AlvanGEdlundCHeddiniAThe global need for effective antibiotics – a summary of plenary presentationsDrug Resist Updat2011142707621440487
- CarletJCollignonPGoldmannDSociety’s failure to protect a precious resource: antibioticsLancet2011378978836937121477855
- KumarasamyKKTolemanMAWalshTREmergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological studyLancet Infect Dis201010959760220705517
- KamatVRNichterMPharmacies, self-medication and pharmaceutical marketing in Bombay, IndiaSoc Sci Med19984767797949690824
- LakshmiVNeed for national/regional guidelines and policies in India to combat antibiotic resistanceIndian J Med Microbiol200826210510718445943
- SahooKCTamhankarAJJohanssonELundborgCSAntibiotic use, resistance development and environmental factors: a qualitative study among healthcare professionals in Orissa, IndiaBMC Public Health20101062920964815
- ShanmugamSSave antibiotics for future of mankindJ Assoc Physicians India201159646521751673
- SubbalaxmiMVLakshmiVLavanyaVAntibiotic resistance – experience in a tertiary care hospital in south IndiaJ Assoc Physicians India201058Suppl182221568007
- VargheseGKMukhopadhyaCBairyIVandanaKEVarmaMBacterial organisms and antimicrobial resistance patternsJ Assoc Physicians India201058Suppl232421563609
- MurrayPR BEPfallerMATenoverFCYolkenRHManual of Clinical Microbiology7th edWashington DCASM Press1999
- Clinical Laboratory Standards InstitutePerformance Standards for Antimicrobial Disk Susceptibility TestsApproved standard, 9th ed CLSI document M2-A9Wayne, PAClinical Laboratory Standards Institute2006
- JarlierVNicolasMHFournierGPhilipponAExtended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patternsRev Infect Dis19881048678783263690
- MagiorakosAPSrinivasanACareyRBMultidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistanceClin Microbiol Infect201218326828121793988
- AshleyEALubellYWhiteNJTurnerPAntimicrobial susceptibility of bacterial isolates from community acquired infections in Sub-Saharan Africa and Asian low and middle income countriesTrop Med Int Health20111691167117921707879
- JeanSSHsuehPRHigh burden of antimicrobial resistance in AsiaInt J Antimicrob Agents201137429129521382699
- BellJMChitsazMTurnidgeJDBartonMWaltersLJJonesRNPrevalence and significance of a negative extended-spectrum beta-lactamase (ESBL) confirmation test result after a positive ESBL screening test result for isolates of Escherichia coli and Klebsiella pneumoniae: results from the SENTRY Asia-Pacific Surveillance ProgramJ Clin Microbiol20074551478148217344367
- BiedenbachDJBellJMSaderHSFritscheTRJonesRNTurnidgeJDAntimicrobial susceptibility of Gram-positive bacterial isolates from the Asia-Pacific region and an in vitro evaluation of the bactericidal activity of daptomycin, vancomycin, and teicoplanin: a SENTRY Program Report (2003–2004)Int J Antimicrob Agents200730214314917531446
- RosenthalVDBijieHMakiDGInternational Nosocomial Infection Control Consortium (INICC) report, data summary of 36 countries, for 2004–2009Am J Infect Control9102011 [Epub ahead of print.]
- BhattacharyaSIs screening patients for antibiotic-resistant bacteria justified in the Indian context?Indian J Med Microbiol201129321321721860099
- MayankDAnshumanMSinghRKAfzalABaroniaAKPrasadKNNosocomial cross-transmission of Pseudomonas aeruginosa between patients in a tertiary intensive care unitIndian J Pathol Microbiol200952450951319805958
- El SolhAAAlhajhusainAUpdate on the treatment of Pseudomonas aeruginosa pneumoniaJ Antimicrob Chemother200964222923819520717
- ZakiSAKarandeSMultidrug-resistant typhoid fever: a reviewJ Infect Dev Ctries20115532433721628808
- Mehnert-KaySADiagnosis and management of uncomplicated urinary tract infectionsAm Fam Physician200572345145616100859
- EshwarappaMDosegowdaRAprameyaIVKhanMWKumarPSKempegowdaPClinico-microbiological profile of urinary tract infection in south IndiaIndian J Nephrol2011211303621655167
- PathakAMahadikKDhaneriaSPSharmaAErikssonBLundborgCSAntibiotic prescribing in outpatients: Hospital and seasonal variations in Ujjain, IndiaScand J Infect Dis2011436–747948821299365
- CastanheiraMDeshpandeLMMathaiDBellJMJonesRNMendesREEarly dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007Antimicrob Agents Chemother20115531274127821189345
- AsensioAAlvarez-EspejoTFernandez-CrehuetJTrends in yearly prevalence of third-generation cephalosporin and fluoroquinolone resistant Enterobacteriaceae infections and antimicrobial use in Spanish hospitals, Spain, 1999 to 2010Euro Surveill20111640 pii 19983