Abstract
Background
Tumor necrosis factor (TNF) family includes lymphotoxin-alpha (LTA) which is a pro-inflammatory cytokine which plays a role in hepatic fibrogenesis. LTA gene polymorphism plays a role in different inflammatory and immunomodulatory diseases. This polymorphism is also suggested to affect chronic hepatitis C (CHC) infection course.
Aim
To study the contribution of LTA gene polymorphism in different chronic hepatitis C stages and hepatocellular carcinoma risk.
Patients and Methods
Our study included 108 chronic HCV patients grouped according to the disease stage. Group (A): CHC, group (B): liver cirrhosis (LC), group (C): LC with HCC, and group (D): healthy controls. Routine laboratory investigations, polymerase chain reaction (PCR) for quantification of HCV, abdominal ultrasonography, and Liver stiffness measurement (LSM) were done. Child–Turcotte–Pugh, Model for end-stage liver disease (MELD), and Fibrosis index based on 4 (FIB-4) scores were calculated. We used the PCR-restriction fragment length polymorphism technique for lymphotoxin-α genotyping.
Results
The A/G genotype was predominant in all groups. In HCC patients, G/G genotype was more frequent (31.8%) than in the LC group (19.4%), CHC group (17.8%), and controls (4.17%). A significant association was found between LTA genotypes and the child classes in HCC (P<0.01) but not in LC patients (P>0.05). HCC patients carrying A/G genotype had higher MELD scores than other genotypes. Multivariate binary logistic regression analysis confirmed that LTA G/G genotype and low platelet count were independent predictors for HCC development in patients with HCV-related LC.
Conclusion
Detection of LTA G/G genotype in chronic HCV patients could help to recognize high-risk patients for disease progression and HCC development.
Introduction
Hepatitis C infection (HCV) is one of the primary causes of chronic liver illness around the world. The estimated anti-HCV antibody seropositivity is 100 million people with viremia in 71 million.Citation1 HCV-related chronic liver diseases cause more than 350,000 deaths every year.Citation2 There is a wide variation in the long-term natural history of the disease ranging from minimal necro-inflammatory changes to extensive fibrosis and cirrhosis with or without hepatocellular carcinoma (HCC).Citation3 Many factors that determine the development of chronic infection with different outcomes are not well recognized, but the role of cytokines and the cellular mediated immunity in the pathogenesis and eradication of chronic HCV have been studied.Citation4,Citation5
Activated macrophages secrete interleukin (IL)-1β and tumor necrosis factor (TNF)-α to promote HCV entry by disrupting hepatocyte tight junctions; however, the mechanism by which this occurred was unrecognized. The TNF superfamily members TNF-α, TNF-β, TWEAK, and LIGHT promote HCV infection through nuclear factor (NF)-κB and myosin light chain kinase (MLCK)-dependent pathway. These data highlight that viral entry and persistent infection may be promoted by HCV-induced TNF superfamily signaling responsesCitation6 and diversity in genetic elements can control and adjust the production of the cytokines.Citation7 The pro-inflammatory cytokines TNF-α and lymphotoxin alpha (previously called TNF-β), are two members of TNF ligand superfamily genes that are encoded by the major histocompatibility complex class III region. Chromosome 6 carries the two genes at near locations. The same receptor is used and stimulation of NF-κB nuclear protein through the same immune-inflammatory responses.Citation8
Lymphotoxin-a (LTA), is a close homolog of TNF-αCitation9 which has about 50% amino acid sequence identity to TNF-α. It carries out most TNF-α activities and both cytokines initiate similar biologic responses.Citation10 LTA is produced in response to T cells stimulation and is involved in different inflammatory, immune-stimulatory, and antiviral responses. It can help communicate lymphocytes and stromal cells and subsequently induce cancer cells cytotoxicity.Citation8,Citation11,Citation12 LTA has been implicated in the pathogenesis of acute and chronic HCV infection.Citation13–Citation15 As an important mediator of hepatic fibrogenesis, TNF-β is released by lymphocytes (T cells, B cells, and natural killer cells) as an inflammatory response in chronic HCV patients.Citation10 Hepatic fibrogenesis is a result of triggering TNF receptor (TNFR) 1 and TNFR2 by TNF-β, inducing the classical and alternative NF-κB signaling pathways.Citation16,Citation17
Several studies examined the role of LT-α polymorphism in different immune-mediated diseases and it has been associated with diseases such as rheumatoid arthritis,Citation18 systemic lupus erythematosus,Citation19 cardiac diseases,Citation20,Citation21 and ankylosing spondylitis.Citation12 Several polymorphisms of the LT-α gene were identified and Messer first reported the existence of a single nucleotide polymorphism (SNP) in the intron of LT-α at position 252 (G>A, designated rs909253, which is associated with overexpression of LT-α.Citation22 As lymphotoxin α is an important mediator of hepatic fibrogenesis, it may be helpful to elucidate the contribution of LTA gene polymorphism in different stages of CHC infection. We aimed to study the association of LTA gene polymorphism +252 (rs909253) with the degree of fibrosis in chronic HCV patients and the severity of HCV-related liver cirrhosis (LC). Also, we aimed to study the contribution of this polymorphism to the HCC risk in chronic HCV-infected patients.
Patients and Methods
Our case–control study was conducted in the Tropical Medicine and Gastroenterology Department, Sohag University Hospital in the period from 2/2016 to 2/2018. One hundred and eight chronic HCV patients in different stages of the disease were included. Chronic HCV infection diagnosis was based on anti-HCV antibodies and HCV RNA positivity. They were grouped based on the disease clinical stage based on clinical, ultrasonographic, and laboratory criteria into three groups. Group A: twenty-eight patients with chronic hepatitis C without cirrhosis. Group B: thirty-six patients with HCV-related liver cirrhosis without HCC. Group C: forty-four patients with HCV-related liver cirrhosis with HCC on top. The presence of hepatic focal lesion(s) on liver ultrasound was confirmed by triphasic computed tomography of the abdomen and/or magnetic resonance imaging (MRI) and serum alpha-fetoprotein measurement to confirm HCC diagnosis. HCC group staging was performed based on Barcelona clinic liver cancer (BCLC) staging.Citation23
Exclusion of patients with hepatitis B co-infection and/or HIV, history suggests autoimmune or drug-induced hepatitis, metastatic hepatic focal lesions, or any hepatic focal lesions other than HCC (benign focal lesion, primary hepatic tumors other than HCC). Also, patients who previously received any treatment modality for HCC and those with extra-hepatic tumors were excluded. Also, 24 apparently healthy sex and age-matched controls were included in the control group (Group D).
Ethical Consideration
All subjects signed a written informed consent at the enrollment time and the Ethical Committee of Sohag Faculty of Medicine approved the study protocol and our study complied with the Declaration of Helsinki.
Methods
All subjects included in the study were subjected to medical history, clinical evaluation, and abdominal ultrasonography. Routine laboratory investigations were done; Complete blood count, liver function tests, serum creatinine, prothrombin time and concentration, and international normalization ratio (INR), and alpha-fetoprotein (AFP).
The following investigations were also done:
Quantification of HCV RNA by PCR: Using Applied biosystems ABI Prism 7000 Sequence detection system, the viral load of HCV RNA was recorded by IU/mL as low, moderate, and high viremia according to the reference values of the laboratory.
Low viremia: <100,000 IU/mL
Moderate viremia: 100,000–1,000,000 IU/mL
High viremia: >1,000,000 IU/mL
Acoustic radiation force impulse (ARFI) elastography was used to measure liver stiffness: Liver stiffness measurement (LSM) was performed using shear wave elastography measured in meters per second (m/sec) (Siemens, syngo S2000 VC25B, AG, Germany). The degree of fibrosis evidenced by PSWE was stratified to the corresponding METAVIR score with the following cutoffs according to Lupsor et al.Citation24
F0 means no scarring (0.8: 1.19 m/sec),
F1 is mild fibrosis (1.2: 1.34 m/sec),
F2 is moderate fibrosis (1.35: 1.60 m/sec)
F3 is severe fibrosis (1.61: 2 m/sec)
F4 is cirrhosis or advanced fibrosis (F4 >2 m/sec).
Child–Turcotte–Pugh (CTP) score: The modified Child score was calculated for LC and HCC groups according to Pugh et al.Citation25 This score depends on encephalopathy, ascites, albumin and levels of bilirubin, and INR. Patients were classified into classes A, B, and C.
Model for end-stage liver disease (MELD) scoreCitation26 was calculated to the cirrhotic and HCC groups. It is based on serum creatinine (mg/dl), total bilirubin (mg/dl), and INR. The following formula was used: MELD = 3.78 × loge (total bilirubin) + 11.2 × loge (INR) + 9.57 × loge (serum creatinine) + 6.43
Fibrosis index based on (FIB-4) score:Citation27 The variables included in the FIB-4 are as simple as age, AST, ALT, and platelet (PLT) count. The FIB-4 index is calculated using the formula: FIB-4 = Age (years) ×AST (U/L)/[PLT (109/L) ×ALT1/2 (U/L)]
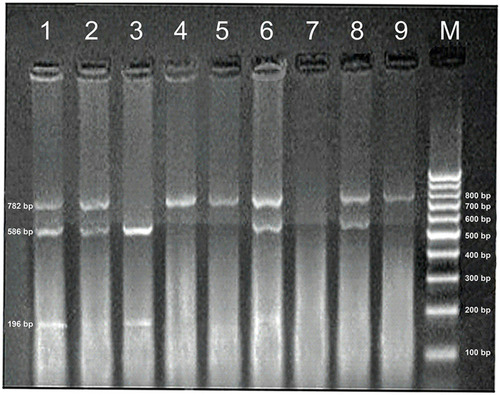
Lymphotoxin-α genotyping by PCR- Restriction Fragment Length Polymorphism (RFLP) technique: Genomic DNA was isolated from collected EDTA preserved blood by buffy coat method using Blood-Plant-Animal DNA Preparation Kits (Cat. No. PP-213S, Jena Bioscience, Germany). For LTA genotyping, we followed the method described previously by Jeng et al, 2014. PCR amplification of the LTA gene was carried out using a forward primer with nucleotide sequence: (5ʹ-CCG TGC TTC GTG CTT TGG ACT A −3ʹ) and a reverse primer with nucleotide sequence: (5ʹ- AGA GGG GTG GTA GCT TGG GTT C-3ʹ). PCR master mix was Ready-to-use (MyTaq Red Mix 2x, Cat. No. BIO-25043, Bioline, London, UK) which contains dNTPs, MgCl2, and enhancers at optimal concentrations. PCR amplification mixture was performed in a final volume of 50 μL containing 2.5 μL of each primer, 25 μL master mix, 5 μL Genomic DNA, and 15 μL deionized water. In each set of experiments, a negative control was included. It was prepared by replacing the DNA template with PCR-grade water. Thermal cycling using a Biometra thermocycler-T Gradient was done. Cycling conditions included one cycle of initial denaturation at 95 °C for 5 min, 35 cycles of denaturation at 94°C for 30 seconds, annealing at 59 °C for 40 seconds and extension at 72°C for 30 seconds, and final extension at 72°C for 10 min. The reaction was stopped by cooling at 4°C. After amplification, the genotypes of LTA +252 (rs909253) were determined by RFLP technique. Digestion of 21.5 µL of the PCR product using 5 units (1 µL) of the fast digest enzyme Nco I (5000 units, Cat. No. EN-E2015-01, Jena Bioscience, Jena, Germany) was done. The digestion mixture also contained 2.5 µL Universal Buffer (UB) 1x and the final volume was 25 μL. The mixture was incubated at 37 °C for 1 hour then visualized by transillumination with UV-light after electrophoresis on a 2% agarose gel and staining with ethidium bromide. The variant G allele contains an Nco I site and is digested into 586 bp and 196 bp fragments. Nco I does not cleave the A allele (782 bp). The heterozygous genotype (A/G) includes the presence of all three fragments. The size of DNA fragments was determined directly by comparison with the 100 bp DNA ladder marker ().
Figure 1 Representative gel picture showing PCR-RFLP analysis of NcoI LTA gene polymorphism on ethidium bromide-stained 2% agarose gel. M stands for molecular size marker (100 bp DNA ladder); lane 3 represents homozygous G/G genotype; lanes 1,2,6,8 represents heterozygous A/G genotype, and lanes 4,5,9 represents homozygous A/A genotype.
Statistical Analysis
Data were analyzed using STATA version 14.2 (Stata Statistical Software: Release 14.2 College Station, TX: StataCorp LP). Quantitative data were represented as mean ± standard deviation, and range. Data were analyzed using Student’s t-test to compare means of two groups and ANOVA for comparison of more than two groups. Qualitative data were presented as numbers and percentages. Either the Chi-square test or Fisher exact test was used for comparison. P-value was considered significant if it was less than 0.05
Results
Demographic data and ultrasonography findings in the included patients are shown in . Splenomegaly was more frequent in LC and HCC groups. Portal vein thrombosis was detected in 13.64% of patients in the HCC group. Ascites were more frequent in patients with liver cirrhosis. Routine laboratory tests revealed thrombocytopenia in all patients groups (CHC, LC, HCC) compared to the controls and in LC and HCC groups compared to the CHC group (P<0.0001). HCC patients had higher serum levels of ALT and AST than LC and CHC groups (P<0.0001 for each). Also, there were significantly lower serum albumin and higher serum bilirubin in both LC and HCC groups compared to the CHC group (P<0.0001 for each). AFP level was significantly higher in HCC than the CHC group (P<0.0001). shows the results of hepatitis C viral load, FIB 4, and LSM determined by ARFI in CHC patients. Most of the patients had low viremia. Most patients (57%) were in F2 and F3 determined by ARFI. Patients in LC and HCC groups were classified according to their Child–Turcotte–Pugh, and MELD scores . Based on the BCLC staging system, the largest proportion of HCC cases was in stage C (38.64%) followed by stage B (34.1%). Less frequently, 20.45% of cases were in stage A and 6.82% in stage D.
Table 1 Demographic Data and Ultrasonography Findings in the Studied Patients
Table 2 The Results of FIB 4, LSM and Hepatitis C Viral Load in CHC Group
Table 3 Child–Turcotte–Pugh and MELD Scores in LC and HCC Groups
Results of PCR Amplification and RFLP Genotyping of LTA Gene +252
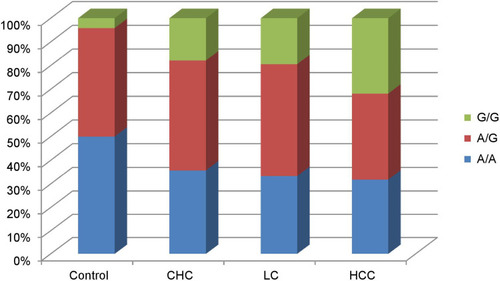
shows the distribution of LTA genotype in patients groups and controls. A/A genotype was the most frequent one in the controls (50%), followed by the A/G genotype and lastly G/G which was the rarest. On the other hand, in all patients groups, the A/G genotype was the predominant variant and A/A genotype was present in about 1/3 of cases and the A/G and G/G were present in nearly 2/3 of patients. In HCC patients, G/G genotype was significantly more frequent (31.8%) than that in LC group (19.4%; P<0.01), CHC group (17.8%; P=0.03) and healthy controls (4.17%; P<0.01).
LTA Genotypes and Other Parameters
In CHC patients, shows no significant association between the three genotypes and LSM by ARFI, FIB 4, and HC viral load. There was a statistically significant association between LTA genotypes and the Child classes in HCC patients (P<0.01) but not in LC patients (P>0.05). We noticed that most HCC patients with A/G genotype were in Child class C and most patients with G/G genotype were in Child B. HCC patients carrying A/G genotype had significantly higher MELD score than other genotypes (P=0.006). However, no statistically significant difference was found between the three genotypes regarding BCLC stages, ultrasonography findings, or AFP levels in HCC patients (P>0.05).
Table 4 The Relation Between LTA Genotypes and Other Parameters
Univariate and Multivariate Logistic Regression Analysis of Factors Predicting HCC
Univariate binary logistic regression analysis revealed that age, platelet count, carriage of LTA G/G genotype, and Child B and C scores were the significant factors associated with HCC development. Multivariate binary logistic regression analysis confirmed that carriage of LTA G/G genotype (OR: 01.09, CI 95%: 1.01–1.40; P=0.01) and platelet count (OR: 0.97, CI 95%: 0.95–0.99; P=0.008) were independent predictors for HCC development in patients with HCV-related LC as shown in .
Table 5 Univariate and Multivariate Binary Logistic Regression Analysis of Predictors of HCC
Discussion
Chronic hepatic inflammation is the hallmark of chronic HCV infection.Citation28,Citation29 Stimulated hepatic Kupffer cells activate nuclear factor-κB to produce pro-inflammatory cytokines including TNF β and TNF α in response to hepatic injuryCitation30 resulting in adverse hepatic fibrosis.Citation17,Citation31 Genetic variations among different individuals can alter the cytokine proteins’ function and this can change the riskCitation32,Citation33 and clinical outcomes of HCC.Citation34 The current study investigates the possible association between the LTA gene polymorphism and severity of HCV-related chronic liver diseases including CHC, liver cirrhosis, and HCC in patients in upper Egypt and its role in disease progression.
In this study, cirrhotic and HCC patients were older than CHC patients. These findings agree with HCV natural history. Mittal and El-SeragCitation35 and ElgamalCitation36 reported that the incidence of HCC increases with age. On other hand, our study demonstrates a male predominance among all groups of chronic liver diseases and it was more prominent in the group (A). Most studies report that HCV infection disproportionately affects males more than females.Citation36–Citation39 Males are known to be at a higher risk of HCC compared with females.Citation40,Citation41 The causes of the difference in HCC risk remain unclear, it may be due to higher prevalence of known risk factors amongst men as HCV infection (eg through injection drug use, tattooing), smokingCitation39 alcohol consumption, higher body mass index, and also increased iron stores.Citation42 One of the suggested explanations for this sexual dimorphism is the protective role of estrogen.Citation40
Our study demonstrates that platelet count was significantly lower in all patients groups (A, B, C) compared to the controls (data not shown). Similarly, both groups (B,C) had significantly lower platelet count compared to the group (A). Thrombocytopenia could be attributed to splenomegaly and hypersplenism secondary to portal hypertension. It may also be due to decreased activity of thrombopoietin.Citation43 Scheiner et alCitation44 found that thrombocytopenia is likely a reflection of HCC phenotypes with different natural histories; where thrombocytopenia is associated with smaller tumor size, less vascular invasion and extrahepatic spread, and better survival compared to patients with normal platelets count or thrombocytosis.
The main causes of morbidity and mortality in chronic liver disease are fibrosis and cirrhosis.Citation17,Citation28 TNF β or lymphotoxin α (LTα) is an important mediator of hepatic fibrogenesisCitation10 and is up-regulated in hepatitis-associated HCC. Different polymorphic sites are present in the LTA gene but there is an SNP at position rs909253 (G>A) in the intron of LTA, which is related to substitution at amino acid position 26 encoded in exon-3 of the LTA gene. It has been shown that over-expression of LTA is due to this polymorphism.Citation8 Our results revealed a predominance of A/A genotype in the control group (50%), followed by the A/G (45.8%) and lastly the G/G variant (4.17%). These results are in accordance with previous studies in Taiwan.Citation31,Citation45
On the other hand, the A/G genotype was the predominant variant in CHC (46.4%) and LC (47.2%) patients in our study. G/G genotype was more predominant in HCC patients (group C) (31.8%) than LC patients (group B) (19.4%; P<0.01), group (A) (17.8%; P=0.03), and healthy controls. These findings are consistent with Jeng et alCitation31 and Tsai et al.Citation45 In the group (A), liver stiffness assessed by ARFI elastography showed that most of the patients were in the F2-F3 stage. We found a significant positive correlation between FIB 4 and LSM using ARFI. The present study did not find any significant association between LTA genotypes and LSM by ARFI, FIB 4, or HCV viral load. Goyal et alCitation13 studied the association of TNF β polymorphism and disease severity in HCV patients based on their histological stage. They concluded that SNP in the TNF β gene might affect the natural course of and the disease progression of chronic HCV infection.
Cagin et alCitation46 found that portal vein thrombosis was associated with severe liver disease and HCC, and it could be a sign of the advanced stage of HCC. Only six (13.6%) of the studied HCC patients had portal vein thrombosis. Gomaa et alCitation47 also reported a comparable frequency and this could be explained by portal vein invasion by malignant thrombus. Regarding LTA genotypes, two of them had A/G and four patients had G/G genotype.
The present study demonstrates that the majority of patients in liver cirrhosis group (B) and HCC group (C) had an advanced stage of the disease according to the Child and MELD scores. In the group (B), we found no significant association between LTA genotype and Child class. However, in HCC patients group (C), a significant association existed. Most patients with A/G genotype were in Child class C and most patients with G/G genotype were in Child class B. Meanwhile, HCC patients carrying A/G genotype had significantly higher MELD scores than those carrying the G/G genotype. When we stratified HCC patients according to the BCLC staging system,Citation23 we found that the largest proportion (39%) was in the advanced stage C, followed by 34% in the intermediate stage B, then 20.5% in the early stage A and a minority (7%) were in the terminal stage D. Moreover, the present study did not find a significant association between both BCLC staging and AFP level, and LTA genotype. Jeng et alCitation31 reported that factors like Child C cirrhosis, thrombocytopenia, and AFP > 400 were associated with carrying TNF β G/G genotype. They also found that this genotype was a biomarker of poor survival of patients with HCC in their study.
Univariate logistic regression analysis of factors predicting the risk of HCC in our study revealed that age, platelet count, LTA genotype, and Child class B and C are significant predictors. However, multivariate analysis identified only platelet count and LTA G/G genotype as independent predictors of HCC in HCV-infected patients. Consistent with our results, Huang et al,Citation48 Everson et al,Citation49 and Yilmaz et alCitation50 stated that platelet count is a well-known risk factor for HCC development in patients with cirrhosis. Thus, follow-up of platelet counts in patients with known cirrhosis can be used to assess the risk of HCC development. Previous studies have shown that increasing age,Citation35,Citation51 male sex,Citation51–Citation53 diabetes mellitusCitation54,Citation55 are independent risk factors for the development of HCC. Jeng et alCitation31 reported independent and additive effects between TNF β G/G genotype and chronic HBV/HCV infection on risk of HCC and this genotype may be used as a biomarker of poor survival. Similarly, Tsai et alCitation45 found independent and additive interactions between TNF β G/G genotype, chronic viral hepatitis, and habits of substance use (as smoking, alcohol, and betel quid chewing) on the risk of HCC.
Our study had certain limitations. Firstly, this study is hospital-based and not a population-based study; therefore, it has a relatively small sample size. Second, all patients and controls were from Upper Egypt (mostly from Sohag Governorate).
In conclusion, we found a significant association between LTA polymorphism (G/G genotype) and HCC development and disease severity in patients with HCV-related liver cirrhosis. LTA G/G genotype and thrombocytopenia may be used as markers to predict the risk of HCC in patients with HCV-related liver cirrhosis. Chronic HCV-infected patients with this variant should receive more intensive screening for early detection of HCC.
Abbreviations
HCV, hepatitis C virus; LTA, lymphotoxin alpha; CHC, chronic hepatitis C; HCC, hepatocellular carcinoma; LC, liver cirrhosis; PCR, polymerase chain reaction; LSM, liver stiffness measurement; MELD, model for end-stage liver disease; FIB-4, fibrosis index based on 4.
Disclosure
The authors report no conflicts of interest in this work.
References
- World Health Organization. Global hepatitis report 2017: web Annex B: WHO estimates of the prevalence and incidence of hepatitis C virus infection by WHO region, 2015.
- World Health Organization. Hepatitis C virus fact sheet, WHO Report 164; 2013. Available from: http://www.who.int/mediacentre/factsheets/fs164/en/in.dex.html. Accessed 58, 2021.
- Pawlotsky JM, Negro F, Aghemo Aet al. EASL (European Association for the Study of the Liver) recommendations on treatment of hepatitis C: final update of the series. J Hepatol. 2020;73:1170–1218.32956768
- Suruki RY, Mueller N, Hayashi K, et al. Host immune status and incidence of hepatocellular carcinoma among subjects infected with hepatitis C virus: a nested case-control study in Japan. Cancer Epidemiol Biomarkers Prev. 2006;15:2521–2525. doi:10.1158/1055-9965.EPI-06-048517164379
- Karbalaei R, Piran M, Rezaei-Tavirani M, AsadzadehAghdaei H, Heidari MH. A systems biology analysis of protein-protein interaction of NASH and IBD based on comprehensive gene information. Gastroenterol Hepatol Bed Bench. 2017;10:194–201.29118935
- Fletcher NF, Clark AR, Balfe P, McKeating JA. TNF superfamily members promote hepatitis C virus entry via an NF-kB and myosin light chain kinase-dependent pathway. J Gen Virol. 2017;98:405–412. doi:10.1099/jgv.0.00068927983476
- Zekri AR, Hafez MM, Bahnassy AA, et al. Genetic profile of Egyptian hepatocellular-carcinoma associated with hepatitis C virus Genotype 4 by 15 K cDNA microarray: a preliminary study. BMC Res Notes. 2008;1:106. doi:10.1186/1756-0500-1-10618959789
- Ghavami SB, Mohebbi SR, Karimi K, et al. Variants in two gene members of the TNF ligand superfamily and hepatitis C virus chronic disease. Gastroenterol Hepatol Bed Bench. 2018;11(Suppl.1):S66–S72.30774809
- Posch PE, Cruz I, Bradshaw D, Medhekar BA. Novel polymorphisms and the definition of promoter ‘alleles’ of the tumor necrosis factor and lymphotoxin α loci: inclusion in HLA haplotypes. Genes Immun. 2003;4(8):547–558. doi:10.1038/sj.gene.636402314647194
- Aggarwal BB, Gupta SC, Kim JH. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey. Blood. 2012;119:651–665. doi:10.1182/blood-2011-04-32522522053109
- Huang Y, Yu X, Wang L, et al. Four genetic polymorphisms of lymphotoxin-alpha gene and cancer risk: a systematic review and meta-analysis. PLoS One. 2013;8(12):e82519. doi:10.1371/journal.pone.008251924349304
- Zhu A, Yang Z, Zhang H, Liu R. Associations of lymphotoxin-a (LTA) rs909253 A/G gene polymorphism, plasma level and risk of ankylosing spondylitis in a Chinese Han population. Sci Rep. 2020;10:1–6. doi:10.1038/s41598-020-57927-631913322
- Goyal A, Kazim SN, Sakhuja P, Malhotra V, Arora N, Sarin SK. Association of TNF‐β polymorphism with disease severity among patients infected with hepatitis C virus. J Med Virol. 2004;72:60–65. doi:10.1002/jmv.1053314635012
- Tsuchiya N, Tokushige K, Yamaguchi N, et al. Influence of TNF gene polymorphism in patients with acute and fulminant hepatitis. J Gastroenterol. 2004;39:859–866. doi:10.1007/s00535-004-1402-115565405
- Suneetha PV, Sarin SK, Goyal A, Kumar GT, Shukla DK, Hissar S. Association between vitamin D receptor, CCR5, TNF-a and TNF-β gene polymorphisms and HBV infection and severity of liver disease. J Hepatol. 2006;44:856–863. doi:10.1016/j.jhep.2006.01.02816545485
- Haybaeck J, Zeller N, Wolf MJ, et al. A lymphotoxin-driven pathway to hepatocellular carcinoma. Cancer Cell. 2009;16:295–308. doi:10.1016/j.ccr.2009.08.02119800575
- Luedde T, Schwabe RFNF. κB in the liver-linking injury, fibrosis, and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2011;8:108–118. doi:10.1038/nrgastro.2010.21321293511
- Newton JL, Harney SJ, Timms AE, et al. Dissection of class III major histocompatibility complex haplotypes associated with rheumatoid arthritis. Arthritis Rheum. 2004;50(7):2122–2129. doi:10.1002/art.2035815248209
- Parks CG, Pandey JP, Dooley MA, et al. Genetic polymorphisms in tumor necrosis factor (TNF)-alpha and TNF-beta in a population-based study of systemic lupus erythematosus: associations and interaction with the interleukin-1alpha-889 C/T polymorphism. Hum Immunol. 2004;65:622–631. doi:10.1016/j.humimm.2004.03.00115219382
- Ozaki K, Ohnishi Y, Iida A, et al. Functional SNPs in the lymphotoxin-α gene that are associated with susceptibility to myocardial infarction. Nat Genet. 2002;32(4):650–654. doi:10.1038/ng104712426569
- Ramasawmy R, Fae KC, Cunha-Neto E, Mullar NG. Polymorphisms in the gene for lymphotoxin-α predispose to chronic Chagas cardiomyopathy. J Infect Dis. 2007;196(12):1836–1843. doi:10.1086/52365318190265
- Messer G, Spengler U, Jung MC, et al. Polymorphic structure of the tumor necrosis factor (TNF) locus. An NcoI polymorphism in the first intron of the human TNF-β gene correlates with a variant amino acid in position 26 and a reduced level of TNF-β production. J Exp Med. 1991;173:209–215. doi:10.1084/jem.173.1.2091670638
- Lovet JM, Brú C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 1999;19:329–338. doi:10.1055/s-2007-100712210518312
- Lupsor M, Badea R, Stefanescu H, et al. Performance of a new elastographic method (ARFI technology) compared to unidimensional transient elastography in the noninvasive assessment of chronic hepatitis C. Preliminary results. J Gastrointestin Liver Dis. 2009;18:303.19795024
- Pugh RN, Murray-Lyon IM, Dawson JL, Pietroni MC, Williams R. Transection of the esophagus for bleeding oesophageal varices. Br J Surg. 1973;60(8):646–649. doi:10.1002/bjs.18006008174541913
- Kamath PS, Wiesner RH, Malinchoc M, et al. A model to predict survival in patients with end-stage liver disease. Hepatology. 2001;33:464–470. doi:10.1053/jhep.2001.2217211172350
- Vallet-Pichard A, Mallet V, Nalpas B, et al. FIB-4: an inexpensive and accurate marker of fibrosis in HCV infection. Comparison with liver biopsy and fibrotest. Hepatology. 2007;46:32–36. doi:10.1002/hep.2166917567829
- El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273. doi:10.1053/j.gastro.2011.12.06122537432
- Su CH, Lin Y, Cai L. Genetic factors, viral infection, other factors, and liver cancer: an update on current progress. Asian Pac J Cancer Prev. 2013;14:4953e60. doi:10.7314/APJCP.2013.14.9.495324175758
- Yang MD, Hsu CM, Chang WS, et al. Tumor necrosis factor-a genotypes are associated with hepatocellular carcinoma risk in Taiwanese males, smokers, and alcohol drinkers. Anticancer Res. 2015;35:5417e23.26408704
- Jeng JE, Wu HF, Tsai MF, et al. Independent and additive interaction between tumor necrosis factor β 252 polymorphisms and chronic hepatitis B and C virus Infection on risk and prognosis of hepatocellular carcinoma: a case-control study. Asian Pac J Cancer Prev. 2014;15:10209–10215. doi:10.7314/APJCP.2014.15.23.1020925556449
- Cheng PN, Chiu HC, Chiu YC, Chen SC, Chen Y. Comparison of FIB-4 and transient elastography in evaluating liver fibrosis of chronic hepatitis C subjects in community. PLoS One. 2018;13:0206947. doi:10.1371/journal.pone.0206947
- Zhang N, Hartig H, Dzhagalov I, Draper D, He YW. The role of apoptosis in the development and function of T lymphocytes. Cell Res. 2005;15:749–769.16246265
- Pan D, Zeng X, Yu H, et al. Role of cytokine gene polymorphisms on prognosis in hepatocellular carcinoma after radical surgery resection. Gene. 2014;544:32–40. doi:10.1016/j.gene.2014.04.04224768182
- Mittal S, El-Serag HB. Epidemiology of HCC: consider the population. J Clin Gastroenterol. 2013;47:S2–S6. doi:10.1097/MCG.0b013e3182872f2923632345
- Elgamal S, Ghafar AA, Ghoneem E, Elshaer M, Alrefai H, Elemshaty W. Characterization of patients with hepatocellular carcinoma on the way for early detection: one center experience. Egypt J Intern Med. 2019;30:231–238. doi:10.4103/ejim.ejim_29_18
- Rantala M, van de Laar MJ. Surveillance and epidemiology of hepatitis B and C in Europe - a review. Euro Surveill. 2008;13:18880. doi:10.2807/ese.13.21.18880-en18761967
- Osório FM, Lauar GM, Lima AS, Vidigal PV, Ferrari TC, Couto CA. Epidemiological aspects of hepatocellular carcinoma in a referral center of Minas Gerais, Brazil. Arq Gastroenterol. 2013;50:97–100. doi:10.1590/S0004-2803201300020001523903617
- Shaker MK, Abdella HM, Khalifa MO, El Dorry AK. Epidemiological characteristics of hepatocellular carcinoma in Egypt: a retrospective analysis of 1313 cases. Liver Int. 2013;33:1601–1606. doi:10.1111/liv.1220923714212
- Liu P, Xie SH, Hu S, et al. Age-specific sex difference in the incidence of hepatocellular carcinoma in the United States. Oncotarget. 2017;8:68131–68137. doi:10.18632/oncotarget.1924528978103
- Lee EY, Xuan Mai TT, Chang Y, Ki M. Trends of liver cancer and its major risk factors in Korea. Epidemiol Health. 2015;37:e2015016. doi:10.4178/epih/e201501625773443
- Nordenstedt H, White DL, El-Serag HB. The changing pattern of epidemiology in hepatocellular carcinoma. Dig Liver Dis. 2010;42:S206–S214. doi:10.1016/S1590-8658(10)60507-520547305
- Afdhal N, McHutchison J, Brown R, et al. Thrombocytopenia associated with chronic liver disease. J Hepatol. 2008;48:1000–1007. doi:10.1016/j.jhep.2008.03.00918433919
- Scheiner B, Kirstein M, Popp S, et al. Association of platelet count and mean platelet volume with overall survival in patients with cirrhosis and unresectable hepatocellular carcinoma. Liver Cancer. 2018;1–15. doi:10.1159/00048714829662829
- Tsai JF, Chen SC, Lin ZY, et al. Interactive effects between Lymphotoxin α +252 polymorphism and habits of substance use on risk of hepatocellular carcinoma. Kaohsiung J Med Sci. 2017;33:334–338. doi:10.1016/j.kjms.2017.04.01028738973
- Cagin YF, Atayan Y, Erdogan MA, Dagtekin F, Colak C. Incidence and clinical presentation of portal vein thrombosis in cirrhotic patients. Hepatobiliary Pancreat Dis Int. 2016;15:499–503. doi:10.1016/S1499-3872(16)60092-927733319
- Gomaa AI, Hashim MS, Waked I. Comparing staging systems for predicting prognosis and survival in patients with hepatocellular carcinoma in Egypt. PLoS One. 2014;9:90929. doi:10.1371/journal.pone.0090929
- Huang YC, Huang CF, Chang KC, et al. Community-based screening for hepatocellular carcinoma in elderly residents in a hepatitis B- and C-endemic area. J Gastroenterol Hepatol. 2011;26:129–134. doi:10.1111/j.1440-1746.2010.06476.x21175806
- Everson GT, Shiffman ML, Hoefs JC, et al.; the HALT-C Trial Group. Quantitative liver function tests improve the prediction of clinical outcomes in chronic hepatitis C: results from the hepatitis C antiviral long-term treatment against cirrhosis trial. Hepatology. 2012;55:1019–1029. doi:10.1002/hep.2475222030902
- Yilmaz Y, Erdal E, Atabey N, Brian IC. Platelets, microenvironment and hepatocellular carcinoma. Biochem Anal Biochem. 2016;5:281.
- Yi SW, Choi JS, Yi JJ, Lee YH, Han KJ. Risk factors for hepatocellular carcinoma by age, sex, and liver disorder status: a prospective cohort study in Korea. Cancer. 2018;124:2748–2757. doi:10.1002/cncr.3140629669170
- McGlynn KA, London WT. The global epidemiology of hepatocellular carcinoma: present and future. Clin Liver Dis. 2011;15:223–243. doi:10.1016/j.cld.2011.03.00621689610
- Sherman M. Primary malignant neoplasms of the liver. In: Dooley JS, Lok AS, Burroughs AK, Heathcote EJ, editors. Sherlock‟s Diseases of the Liver and Biliary System. 12th ed. Oxford: Blackwell Publishing Ltd; 2011:681–703.
- Wang P, Kang D, Cao W, Wang Y, Liu Z. Diabetes mellitus and risk of hepatocellular carcinoma: a systematic review and meta-analysis. Diabetes Metab Res. 2012;28:109–122. doi:10.1002/dmrr.1291
- Tang A, Hallouch O, Chernyak V, Kamaya A, Sirlin CB. Epidemiology of hepatocellular carcinoma: target population for surveillance and diagnosis. Abdom Radiol. 2018;43:13–25. doi:10.1007/s00261-017-1209-1