Abstract
Background
Vibrio vulnificus (VV) is a causative agent of foodborne diseases with high mortality. The aim of this study was to investigate the genomic and phenotypic profiles of VV.
Methods
Six VV isolates were collected and conducted whole-genome sequencing. Biofilm formation and anti-complement killing test were performed to evaluate the pathogenicity. Subsequently, 157 publicly available genomes of VV isolates were selected to determine the evolutionary relationship.
Results
The resistant genes norM and tet34 were identified in six isolates. A total of 156 virulence genes were identified. However, there is no obvious difference between strains isolated from blood and puncture fluid. The tendency of growth for six isolates decreased with the lapse of time, while the biofilm formation increased. The genes tadC and flp related to Flp pili were found in isolate 25506 and 30896, resulting in more obvious biofilm formation. In addition, the survival rate of 19656 was less than 20% due to lack of one genomic island including virulence genes (impD-H, clpV-1) relevant to type VI secretion system (T6SS). Multi-locus sequence typing (MLST) revealed 95 different STs and 19 novel STs, indicating that the tendency of 163 isolates was sporadic. Further comparative genomics analysis clearly classified 163 isolates into three distinct evolutionary lineages.
Conclusion
VV infections were sporadic in humans and the environment. Virulence genes impD-H and clpV-1 related to T6SS were associated with pathogenicity phenotype of VV.
Background
Fisheries and aquaculture are becoming increasingly intensive to meet recent human consumption, resulting in proliferation of marine pathogens and food security concerns.Citation1,Citation2 Vibrio species, as one of the most dangerous foodborne pathogens, cause vibriosis in human around the world.Citation3 It has been reported that vibriosis resulted in 80,000 illnesses and 100 deaths each year in the United States.Citation4 Among Vibrio species, V. vulnificus (VV) is responsible for multiple outbreaks in fish farms and for the highest fatality rate in human.Citation4–Citation6 Previous study demonstrated VV could activate genes involved in colonization and resistance in the blood, correlating with global warming.Citation7 Surveillance data from the Cholera and Other Vibrio Illness Surveillance (COVIS) system indicated an increase tendency of VV infections in recent years in the United States.Citation4
In China, VV infections are very rare and there are no reported outbreaks in human.Citation8 Epidemiological investigations showed an overlap in the distribution of VV among marine animals, aquariums, and human.Citation9,Citation10 However, the genetic factors that differentiate clinical or environmental isolates remain enigmatic. Pangenome analysis has identified VV isolates cohabitated with both commensal and bloomer ecotypes in the mucosa of eels from aquaculture farms, raising concerns about man-made environments.Citation11 The global overview of VV epidemiology is still limited partly due to imperfect surveillance systems. Therefore, the aim of this study was to investigate the genomic and phenotypic profiles of VV isolated from our hospital and gain insights into the evolutionary differences of VV in human and non-human.
Methods
Collection of Bacterial Strains and Whole-Genome Sequencing (WGS)
Six VV isolates (5955, 19656, 25506, 30896, 32999, 41678) were collected from patients as described previously.Citation12 Genomic DNA was extracted by FastDNA SPIN Kit for Soil (MP Biomedicals, United States) and sequenced using HiSeq 2000 (Illumina, SanDiego, CA, USA).
All genomes were annotated using Prokka.Citation13 The analysis of pathogenicity, virulence genes, resistance genes and genomic island was performed as described previously.Citation14
The Whole Genome Shotgun BioProject for the six VV isolates has been deposited under BioProject accession No. PRJNA714541.
Growth Curve and Biofilm Formation Testing
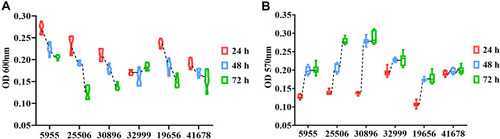
Six isolates were inoculated into 96-well polystyrene microtiter plates containing 200 μL Mueller–Hinton broth (MHB) for 24 h, 48 h, and 72 h. After static incubation, the absorbance was tested using a plate reader at 600 nm. Then, the biofilm formation was further tested as described previously.Citation15
Anti-Complement Killing Test
Mouse serum was obtained from Guangzhou Ruite Biotechnology (Guangzhou, China). The serum was placed in a water bath at 56°C for 30 min to inactivate complement, generating inactive serum. The overnight bacterial culture was diluted to a cell density of 2×106 CFU/mL, and normal and inactivated sera (180 μL) were separately mixed with 20 μL bacterial suspension and incubated at 37°C for 1 h. Samples were diluted 100-fold and spread onto plates. After overnight incubation, the colonies on plates were counted. The bacterial survival rate was calculated using the following formula:
Bacterial survival rate = (number of colonies with normal serum/number of colonies with inactivated serum) × 100%.
Comparative Genomic Analysis
Six genomes in the present study and 157 publicly available VV genomes were selected to determine the evolutionary relationship (Supplementary Table 1). Multi-locus sequence typing (MLST) analysis used an online tool (http://www.genomicepidemiology.org/). Core genome for the genome dataset was calculated by Roary (https://sanger-pathogens.github.io/Roary/). The resulting consensus tree was visualized and edited using the Interactive Tree of Life (iTOL).Citation16
Results
Antibiotic Resistance Mechanism of VV
The antimicrobial susceptibility testing of six isolates was performed in a previous study.Citation12 Except polymyxin B and gentamicin, six VV isolates were sensitive to other tested antibiotics. The resistant genes norM and tet34 were identified in all six included isolates, inducing efflux pump and tetracycline resistance, respectively.
Virulence Genes
A total of 156 virulence genes were identified, and 133 genes were found in all six isolates (Supplementary Table 2). These genes are possibly associated with pathogenicity, including capsular polysaccharide, flagella, flp pili, multifunctional-auto processing repeats-in-toxin (MARTX) toxin, hemolysin, mannose-sensitive hemagglutinin, and two secretion systems (type II and VI). There is no obvious difference between strains isolated from blood and puncture fluid.
Characteristics of Pathogenicity
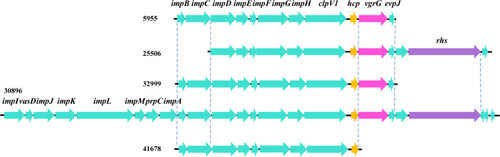
As shown in , the growth of six isolates diminished with the lapse of time. However, the tendency of biofilm formation increased with the time in all isolates, especially for isolates 25506 and 30896 (). The genes (tadC and flp) related to Flp pili were found only in isolates 25506 and 30896. In addition, the survival rate of 19656 was less than 20% (Supplemental Figure 1). Notably, except 19656, other five isolates had one genomic island including virulence genes (impD-H, clpV-1) related to type VI secretion system (T6SS) ().
Comparative Genomics Analysis
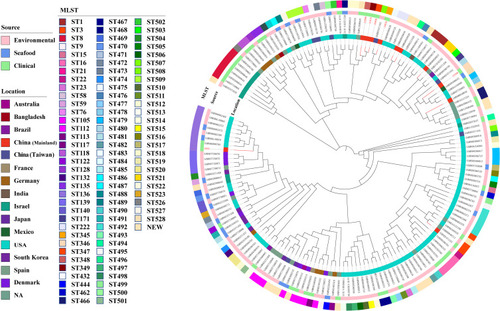
The data of our genome collections showed the incidence of VV infections was highest in the United States. There were 95 different STs and 19 novel STs among the 163 isolates. The most common STs were ST136 (8, 4.9%), ST112 (7, 4.3%), and ST8 (6, 3.7%). Therefore, the epidemic of VV infections had sporadic tendency. Comparative genomics analysis based on core genome clearly classified 163 isolates into three distinct evolutionary lineages with most isolates grouped into lineages III (). Seven and fifty-six isolates detected in this study were grouped into lineages I and II, respectively. Isolates in lineages I were only collected from environment and clinic. The proportion of strains isolated from human in lineages II (16, 28.6%) was higher than that in lineages III (20, 20%).
Figure 3 Core-genome-based phylogenetic tree of 163 VV isolates, including 6 isolates from this study and 157 strains downloaded from NCBI genome database. STs of the isolates is labelled in the outer ring. The source of all isolates is presented in the middle ring. The location of the isolates is colored in the inner ring. The number of isolates in the present study was colored in red.
Discussion
VV is a zoonotic pathogen, posing a threat to marine animals, aquariums and humans.Citation1,Citation3–Citation5 Here, a positive correlation was demonstrated between the expression of genes tadC and flp and biofilm formation of VV. In addition, virulence genes (impD-H and clpV-1) related to T6SS exhibited association with survival rate in the serum. Furthermore, VV from the world displayed a divergent population structure with extensive diversification based on comparative genomics.
In the present study, a large and geographically diverse strains were included, encompassing environmental, seafood and clinical strains. MLST revealed VV infections appeared sporadic. Comparative genomics analysis based on core genome clearly classified all isolates into three distinct evolutionary lineages. The vast majority of cases occurred in the United States. The possible reason is that few countries, with the exception of the United States, maintain dedicated and legally enforced surveillance systems for Vibrio species.Citation17 With an increase in the incidence of VV infections, especially for those emerging with global warming, more attention should be paid to systematical monitoring of VV in different sources and locations.Citation3
The pathogenesis of VV is multi-faceted, but poorly understood. A wide array of putative virulence factors were involved in VV possess, such as acid neutralization, capsular polysaccharide, iron acquisition, cytotoxicity systems, motility, and proteins related to attachment and adhesion.Citation3,Citation18 Unfortunately, no single virulence gene has been identified as a critical factor for human virulence. Similarly, we found no obvious difference between strains isolated from blood and puncture fluid based on genomic analysis. Therefore, the hunt for specific molecular markers that could distinguish pathogenic and non-pathogenic VV needs to be continued.
Biofilms are three-dimensional complex matrix structures that might favor cross-contamination of aquatic organisms, such as Vibrio species.Citation19 Previous studies demonstrated that biofilm formation is important for the ecology, transmission and virulence of VV and V. parahaemolyticus.Citation20,Citation21 The isolates variability could lead to the heterogeneous biofilm formation.Citation22,Citation23 The tendency of biofilm formation increased with the time in all six VV isolates, especially for isolates 25506 and 30896 with genes tadC and flp. As previous study noted, the tad operons encode the machinery required for adhesive Flp pili biogenesis for VV, resulting in increased biofilm formation, auto-aggregation, and oyster colonization.Citation24 In addition, Pu et al found deletion of flp altered the near-surface motility profile of VV that diminished bacteria-surface interactions.Citation25 However, this area of research remains underdeveloped. Therefore, understanding the roles of Flp pili for motility during biofilm development remains an important area of investigation.
T6SS, as a molecular syringe composed of 13 essential proteins, plays an important role in the injection of cytotoxic effectors into neighbouring cells.Citation26 T6SS has been implicated in both anti-prokaryotic and anti-eukaryotic activity as well.Citation27,Citation28 The isolate 19656 was lacking one genomic island including virulence genes (impD-H, clpV-1) related to T6SS, causing the survival rate to be less than 20% in the anti-complement killing test. These findings suggest that the T6SS may be good potential to serve as a predictive index of pathogenic VV in clinical infections. Further studies to evaluate this possibility are warranted.
This study has several limitations. First, the included sample size for phenotypic identification was small. However, a system evaluation for evolutionary relationship was performed in the present study. In addition, the pathogenicity of VV isolates was not further confirmed by animal experiments. Therefore, a further large-scale study is needed for better evaluation of specific epidemiological factors and potential significance of virulence genes for driving VV infections.
Conclusions
In conclusion, VV infections occurred as sporadic cases. Virulence genes (impD-H, clpV-1) related to T6SS were associated with pathogenicity phenotype of VV. In addition, future investigations involving more human and non-human isolates will be required to identify reliable correlation between virulence genes and observed virulence.
Data Sharing Statement
The sequencing data for VV has been deposited at GenBank under BioProject accession No. PRJNA714541.
Ethics Approval
We declare no ethical competing interest. In our study, we did not perform any experiments with animals or higher invertebrates, neither performed experiments on humans nor the use of human tissue samples. Our data have been originated from bacteria, not linked to clinical information.
Consent for Publication
All authors have seen and approved the content and fulfil the journal’s requirements for authorship.
Disclosure
The authors report no conflicts of interest in this work.
References
- GräslundS, BengtssonBE. Chemicals and biological products used in south-east Asian shrimp farming, and their potential impact on the environment–a review. Sci Total Environ. 2001;280(1–3):93–131. doi:10.1016/s0048-9697(01)00818-x11763276
- HubertCL, MichellSL. A universal oyster infection model demonstrates that Vibrio vulnificus type 6 secretion systems have antibacterial activity in vivo. Environ Microbiol. 2020;22(10):4381–4393. doi:10.1111/1462-2920.1512332515159
- Baker-AustinC, OliverJD. Vibrio vulnificus: new insights into a deadly opportunistic pathogen. Environ Microbiol. 2018;20(2):423–430. doi:10.1111/1462-2920.1395529027375
- Centers for Disease Control and Prevention. Cholera and Other Vibrio Illness Surveillance (COVIS). Available from: https://www.cdc.gov/nationalsurveillance/cholera_vibrio_surveillance.htm. Accessed March 05, 2019.
- KasparCW, TamplinML. Effects of temperature and salinity on the survival of Vibrio vulnificus in seawater and shellfish. Appl Environ Microbiol. 1993;59(8):2425–2429. doi:10.1128/AEM.59.8.2425-2429.19938368832
- TaoZ, LarsenAM, BullardSA, WrightAC, AriasCR. Prevalence and population structure of Vibrio vulnificus on fishes from the northern Gulf of Mexico. Appl Environ Microbiol. 2012;78(21):7611–7618. doi:10.1128/AEM.01646-1222923394
- Hernández-CabanyeroC, AmaroC. Phylogeny and life cycle of the zoonotic pathogen Vibrio vulnificus. Environ Microbiol. 2020;22(10):4133–4148. doi:10.1111/1462-2920.1513732567215
- ChuangYC, YuanCY, LiuCY, LanCK, HuangAH. Vibrio vulnificus infection in Taiwan: report of 28 cases and review of clinical manifestations and treatment. Clin Infect Dis. 1992;15(2):271–276. doi:10.1093/clinids/15.2.2711520762
- LiM, ZhaoL, MaJ, et al. Vibrio vulnificus in aquariums is a novel threat to marine mammals and public health. Transbound Emerg Dis. 2018;65(6):1863–1871. doi:10.1111/tbed.1296730047566
- GibelloA, VelaAI, Martínez-NevadoE, et al. Potentially human-virulent Vibrio vulnificus isolates from diseased great pompano (Trachinotus goodei). Transbound Emerg Dis. 2019;66(4):1765–1770. doi:10.1111/tbed.1319030927558
- López-PérezM, JayakumarJM, Haro-MorenoJM, et al. Evolutionary model of cluster divergence of the emergent marine pathogen Vibrio vulnificus: from genotype to ecotype. mBio. 2019;10(1):e02852–18. doi:10.1128/mBio.02852-1830782660
- YuW, ShenX, PanH, XiaoT, ShenP, XiaoY. Clinical features and treatment of patients with Vibrio vulnificus infection. Int J Infect Dis. 2017;59:1–6. doi:10.1016/j.ijid.2017.03.01728347850
- SeemannT. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi:10.1093/bioinformatics/btu15324642063
- YuW, HuangY, YingC, et al. Analysis of genetic diversity and antibiotic options for clinical listeria monocytogenes infections in China. Open Forum Infect Dis. 2021;8(6):ofab177. doi:10.1093/ofid/ofab17734159214
- YuW, ZhangJ, TongJ, et al. In vitro antimicrobial activity of fosfomycin, vancomycin and daptomycin alone, and in combination, against linezolid-resistant Enterococcus faecalis. Infect Dis Ther. 2020;9(4):927–934. doi:10.1007/s40121-020-00342-132964392
- LetunicI, BorkP. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. doi:10.1093/nar/gkz23930931475
- NewtonA, KendallM, VugiaDJ, HenaoOL, MahonBE. Increasing rates of vibriosis in the United States, 1996–2010: review of surveillance data from 2 systems. Clin Infect Dis. 2012;54(Suppl 5):S391–5. doi:10.1093/cid/cis24322572659
- JonesMK, OliverJD. Vibrio vulnificus: disease and pathogenesis. Infect Immun. 2009;77(5):1723–1733. doi:10.1128/IAI.01046-0819255188
- Lucero-MejíaJE, Romero-GómezSJ, Hernández-IturriagaM. A new classification criterion for the biofilm formation index: a study of the biofilm dynamics of pathogenic Vibrio species isolated from seafood and food contact surfaces. J Food Sci. 2020;85(8):2491–2497. doi:10.1111/1750-3841.1532532654171
- GuligPA, BourdageKL, StarksAM. Molecular pathogenesis of Vibrio vulnificus. J Microbiol. 2005;43:118–131.15765065
- YildizFH, VisickKL. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 2009;17(3):109–118. doi:10.1016/j.tim.2008.12.00419231189
- MizanMF, JahidIK, HaSD. Microbial biofilms in seafood: a food-hygiene challenge. Food Microbiol. 2015;49:41–55. doi:10.1016/j.fm.2015.01.00925846914
- BeshiruA, IgbinosaEO. Characterization of extracellular virulence properties and biofilm-formation capacity of Vibrio species recovered from ready-to-eat (RTE) shrimps. Microb Pathog. 2018;119:93–102. doi:10.1016/j.micpath.2018.04.01529654902
- Duong-NuTM, JeongK, HongSH, et al. A stealth adhesion factor contributes to Vibrio vulnificus pathogenicity: flp pili play roles in host invasion, survival in the blood stream and resistance to complement activation. PLoS Pathog. 2019;15(8):e1007767. doi:10.1371/journal.ppat.100776731437245
- PuM, Rowe-MagnusDA. A Tad pilus promotes the establishment and resistance of Vibrio vulnificus biofilms to mechanical clearance. NPJ Biofilms Microbiomes. 2018;4(1):10. doi:10.1038/s41522-018-0052-729707230
- HoBT, DongTG, MekalanosJJ. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe. 2014;15(1):9–21. doi:10.1016/j.chom.2013.11.00824332978
- TrunkK, PeltierJ, LiuYC, et al. The type VI secretion system deploys antifungal effectors against microbial competitors. Nat Microbiol. 2018;3(8):920–931. doi:10.1038/s41564-018-0191-x30038307
- RayA, SchwartzN, de Souza SantosM, ZhangJ, OrthK, SalomonD. Type VI secretion system MIX-effectors carry both antibacterial and anti-eukaryotic activities. EMBO Rep. 2017;18(11):1978–1990. doi:10.15252/embr.20174422628912123