Abstract
Purpose
To establish a typing scheme for IncFIB replicon and to dissect genomic features of IncFIB-4.1/4.2 single-replicon plasmids.
Methods
A total of 146 representative fully sequenced IncFIB-replicon-containing plasmids were selected to construct a phylogenetic tree of repBIncFIB sequences. A collection of nine IncFIB-4.1/4.2 single-replicon plasmids from China were fully sequenced here and compared with the first sequenced IncFIB-4.1/4.2 single-replicon plasmids from GenBank to dissect their genomic diversity.
Results
In this study, a repB sequence-based scheme was proposed for grouping IncFIB replicon into seven primary types and further into 70 subtypes. A collection of nine IncFIB-4.1/4.2 single-replicon plasmids were fully sequenced here and compared with the first sequenced IncFIB-4.1/4.2 single-replicon plasmids from GenBank. These 11 plasmids had small backbones and shared only three key backbone markers repB together with its iterons, parABC, and stbD. Each plasmid contained one large accessory region (LAR) inserted into the backbone, and these 11 LARs had significantly distinct profiles of mobile genetic elements (MGEs) and resistance/metabolism gene loci. Antibiotic resistance regions (ARRs; the antibiotic resistance gene-containing genetic elements) were found in seven of these 11 LARs. Besides resistance genes, ARRs carried unit or composite transposons, integrons, and putative resistance units. IncFIB-4.1/4.2 single-replicon plasmids were important vectors of drug resistance genes. This was the first report of three novel MGEs: In1776, Tn6755, and Tn6857.
Conclusion
Data presented here provided a deeper insight into diversity and evolution of IncFIB replicon and IncFIB-4.1/4.2 single-replicon plasmids.
Introduction
IncF plasmids, initially found in Escherichia coli K-12 in 1946,Citation1 are widely spreading among Escherichia coli,Citation2 Klebsiella pneumonia,Citation3 Salmonella enterica,Citation4 et al. They acted as vectors of diverse mobile genetic elements (MGEs) carrying various resistance,Citation5–7 virulenceCitation8 or metabolism-related genes.Citation9,Citation10 By the intercellular transfer of IncF plasmids, these genes were accumulated and disseminated in different bacterial isolates, and thus enhances the adaption and survival of these IncF plasmid-containing isolates.
Based on replication initiation protein Rep sequences, different F-type replicons have been identified and can be further categorized into IncFII, IncFIA, and IncFIB.Citation11 IncFII are regulated by antisense RNAs, while IncFIA and IncFIB function as iteron-regulated replicons.Citation12,Citation13 IncFIA and IncFIB often as auxiliary replicons coexist with the major IncFII replicon to constitute multi-replicon plasmids, as represented by the IncFII:IncFIA plasmid p28078-NDM (GenBank accession number MF156713), the IncFII:IncFIB plasmid pO86A1 (GenBank accession number AB255435), and the IncFII:IncFIA:IncFIB plasmid pCA08.Citation14 Additionally, other replicons, such as IncR and IncN, can also cointegrate with IncFII, as represented by the IncFII:IncFIB:IncR plasmid pKPHS2Citation15 and the IncFII:IncpA1763-KPC:IncN1 plasmid pA3295-KPC.Citation16 This multi-replicon status will overcome the IncF incompatibility barrier and accomplish the broad host range, thereby facilitating dissemination of IncF plasmids.Citation17 IncFIB as a sole replicon or an auxiliary replicon in a plasmid is also capable of guaranteeing the stable maintenance for inheritance, as presented by the plasmid pSE11-3Citation18 or Plasmid F.Citation12
There are few reports on characterization of IncFIB repliconCitation11 and IncFIB single-replicon plasmids.Citation19 In this work, a repB sequence-based scheme was proposed for grouping IncFIB replicon into seven primary types and further into 70 subtypes. Furthermore, a comprehensive comparative genomics analysis of 11 IncFIB-4.1/4.2 single-replicon plasmids (including nine ones sequenced in this work) provided a deeper insight into diversification and evolution of IncFIB-4.1/4.2 single-replicon plasmids.
Materials and Methods
Bacterial Strains and Identification
Nine clinical isolates, including Klebsiella pneumoniae BJ20, 71221, W08291, 10057 and A2359, K. quasipneumoniae A2508, 13294 and A1876, and Leclercia adcarboxglata L21, were collected from eight different Chinese public hospitals (Table S1). Each of them carried an IncFIB single-replicon plasmid that designated as pBJ20-tetA, p71221-mphA, pW08291-tetA, p10057-catA, pA2359-IMP, pA2508-emrE, p13294-1NR, pA1876-NR, and pL21-1NR, respectively (Table S2). These nine plasmids had complex molecular structures and contained multiple loci related to antibiotics, heavy metal, or metabolism resistance. To provide a comprehensive comparative genomics analysis of IncFIB-4.1/4.2 plasmids, these nine plasmids were involved into a comprehensive comparative genomics analysis.
Genomic DNA Extraction, Sequencing, and Sequence Assembly
Bacterial genomic DNA was isolated using the UltraClean Microbial Kit (Qiagen, NW, Germany), and sequenced from a sheared DNA library with average size of 15 kb (ranged from 10 kb to 20 kb) on a PacBio RSII sequencer (Pacific Biosciences, CA, USA),Citation20 as well as a paired-end library with an average insert size of 350 bp (ranged from 150 bp to 600 bp) on a HiSeq sequencer (Illumina, CA, USA). The paired-end short Illumina reads were used to correct the long PacBio reads utilizing proovread,Citation21 and then the corrected PacBio reads were assembled de novo utilizing SMARdenovo (https://github.com/ruanjue/smartdenovo).
Sequence Annotation and Comparison
Genome sequences were annotated by the Rapid Annotation using Subsystem Technology (RAST)Citation22 combined with BLASTP/BLASTX/BLASTN,Citation23 Domain (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), and RefSeq database.Citation24 Annotation of replication genes, resistance genes, MGEs, and other features were carried out using the online databases including PlasmidFinder,Citation25 CARD,Citation26 ResFinder,Citation27 ISfinder,Citation28 INTEGRALL,Citation29 and Tn Number Registry.Citation30 Multiple and pairwise sequence comparisons were performed using BLASTN. Gene organization diagrams were drawn using Inkscape 1.0 (https://inkscape.org/en/).
Phylogenomic Analysis
The repBIncFIB sequences of indicated plasmids were aligned using Clustal Omega 1.2.2,Citation31 and then maximum-likelihood phylogenetic trees were constructed from aligned sequences using MEGA X 10.1.8Citation32 with a bootstrap iteration of 1000.
Conjugal Transfer and Electroporation Transfer
Each indicated plasmid was transformed from its wild-type isolate into E. coli EC600 or TOP10 in Enterobacteriaceae, through conjugal transfer or electroporation experiments, respectively. Three milliliters of overnight cultures of each donor and recipient bacteria were mixed together, harvested, and resuspended in 80 mL of Brain Heart Infusion (BHI) broth (BD Biosciences). The mixture was spotted on a 1 cm2 hydrophilic nylon membrane filter with a 0.45 µm pore size (Millipore) that was placed on BHI agar (BD Biosciences) plate and then incubated for mating at 26°C or 37°C for 12 h to 18 h. Bacteria were washed from filter membrane and spotted on BHI plates, for selecting mphA-carrying transconjugant and catA-carrying transconjugant. 1500 μg/mL rifampin (for EC600), together with 40 μg/mL azithromycin [for mph(A)] or 25 μg/mL chloramphenicol (for catA) was used as transconjugant selection, respectively.
To prepare competent cells for electroporation, 200 mL of overnight culture of E. coli TOP10 in Super Optimal Broth (SOB) at an optical density (OD600) of 0.4 to 0.6 was washed three times with electroporation buffer (0.5 M mannitol and 10% glycerol) and concentrated into a final volume of 2 mL. One microgram of DNA was mixed with 100 μL of competent cells for electroporation at 25 μF, 200 Ω, and 2.5 Kv. The resulting cells were suspended in 500 μL of SOB, and an appropriate aliquot was spotted on SOB agar plates containing 40 μg/mL azithromycin [for mph(A)] or 25 μg/mL chloramphenicol (for catA), for selecting of E. coli electroporant, respectively.
Bacterial Antimicrobial Susceptibility Testing
Bacterial antimicrobial susceptibility was tested by E-test, and bacterial antimicrobial susceptibility was interpreted as per the 2020 Clinical and Laboratory Standards Institute (CLSI) guidelines.Citation33
Nucleotide Sequence Accession Numbers
Complete sequences of plasmids pBJ20-tetA (K. pneumoniae BJ20), p71221-mphA (K. pneumoniae 71221), pW08291-tetA (K. pneumoniae W08291), pA2508-emrE (K. quasipneumoniae A2508), pL21-1NR (L. adcarboxglata L21), p10057-catA (K. pneumoniae 10057), p13294-1NR (K. quasipneumoniae 13294), pA1876-NR (K. quasipneumoniae A1876), and pA2359-IMP (K. pneumoniae A2359) were submitted to GenBank under accession numbers MN310373, MN310374, MN310376, MN310379, MN423365, MN423364, MT570100, MT549899, and MN423363, respectively.
Results and Discussion
A Typing Scheme for repBIncFIB Genes
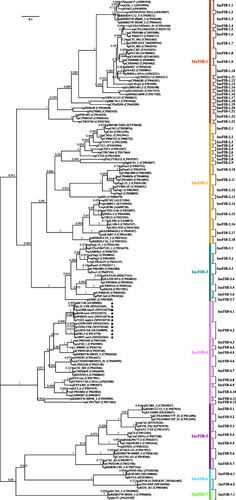
A phylogenetic tree () was constructed from the repBIncFIB sequences of 146 arbitrarily selected representative plasmids (Table S3; last accessed November 2nd 2020) with IncFIB as the sole or auxiliary replicon. repBIncFIB could be divided into seven major separately clustering clades ( and S1), being designated as primary IncFIB types IncFIB-1 to IncFIB-7. Each primary type was further divided into various subtypes (eg IncFIB-1.1 to IncFIB-1.23), which were based on the criterion that repBIncFIB sequences within each subtype displayed ≥95% nucleotide identity while <95% sequence identity was observed between different subtypes. Accordingly, a total of 70 IncFIB subtypes were identified.
Figure 1 A maximum-likelihood phylogenetic tree of repBIncFIB sequences.
A Group of 11 IncFIB-4 Single-Replicon Plasmids Analyzed
Five IncFIB-4.1 single-replicon plasmids and four IncFIB-4.2 single-replicon plasmids were fully sequenced here. A genomic comparison was applied to a total 11 plasmids (Table S2), which comprised the above nine ones and also two GenBank plasmids pSC138 and pFB2.3, which were the first sequenced IncFIB-4.1/4.2 single-replicon plasmids and had the minimum IncFIB-4.1/4.2 backbones, respectively.
The modular structure of each plasmid was divided into the backbone, and the accessory modules that manifested as acquired DNA regions associated/bordered with MGEs (Figure S2 and Table S4). These 11 plasmids had small backbones (6.9 kb to 24.3 kb in length) and contained no conjugal transfer genes. They shared only three key backbone markers repB together with its iterons (replication), parABC (partition), and stbD (mediator of plasmid stability), but displaying remarkable sequence divergence between IncFIB-4.1 and IncFIB-4.2. Each plasmid carried one large accessory region (LAR): the resulting 11 LARs varied in size from about 98 kb to nearly 236 kb and had significantly different profiles of MGEs and resistance/metabolism genes. At least 46 resistance genes, involved in resistance to 17 categories of antibiotics and heavy metals, were identified in these 11 plasmids (Table S5).
The six IncFIB-4.1 plasmids displayed >96% nucleotide identity across 25% to 100% of their backbone sequences, while the five IncFIB-4.2 plasmids showed ≥99% nucleotide identity over ≥84% of their backbone sequences; by contrast, IncFIB-4.1 and IncFIB-4.1 plasmids had ≤90% nucleotide identity across ≤90% of their backbone sequences (Table S6).
Modular Differences in IncFIB-4.1 Single-Replicon Plasmids
Based on a presumed prototype IncFIB-4.1 backbone, the six IncFIB-4.1 plasmids exhibited at least eight major modular differences (Figure S2): i) integration of two copies of IS903B led to a 5.1-kb backbone deletion and a 5.7-kb backbone inversion in pBJ20-tetA, respectively; ii) ISKpn26 was inserted at two different sites of the backbone in pBJ20-tetA and pW08291-tetA; iii) insertion of IS1A–ISSen4 led to absence of the backbone gene orf306 in pBJ20-tetA, p71221-mphA, and pW08291-tetA; iv) insertion of IS903B caused a 3.8-kb backbone deletion in p71221-mphA and pW08291-tetA; v) a group II Kl.pn.I5-like intron was integrated into p71221-mphA, pW08291-tetA, and pBJ20-tetA; vi) LAR integration resulted in 14.7-kb and 2.6-kb backbone deletions upstream and downstream, respectively, of LAR in both pL21-1NR and pSC138, and led to a 3.8-kb upstream-of-LAR deletion in pA2508-emrE; vii) there was the deletion of a 4.2-kb orf1953–to–orf426 backbone region from pA2508-emrE, pL21-1NR, and pSC138; viii) the copy numbers of 43-bp tandem repeat within parC (ParB-binding sites) varied as 8, 8, 8, 19, 11, and 11 in pBJ20-tetA, p71221-mphA, pW08291-tetA, pA2508-emrE, pL21-1NR, and pSC138, respectively. Together, IncFIB-4.1 backbones underwent massive deletion events, mostly resulting from accessory module integration.
The six LARs (Figure S2) had at least six major genetic differences. Firstly, antibiotic resistance regions (ARRs; the antibiotic resistance gene-containing genetic elements in LARs) were identified in pBJ20-tetA, p71221-mphA, pW08291-tetA, pA2508-emrE, and pSC138. Secondly, ars loci (arsenic resistance) were found in pBJ20-tetA, p71221-mphA, pW08291-tetA, pA2508-emrE, and pL21-1NR; Thirdly, sil–cop regions, conferring resistance to silver (sil) and copper (cop), were present in the five plasmids except for pSC138. Fourthly, pA2508-emrE had five metabolism gene loci: phn (phosphonate uptake), urt (urea uptake), fec (iron uptake), lac (galactoside uptake), and gsi (glutathione uptake). Fifthly, a K-antigen gene cluster was present only in pL21-1NR. Sixthly, a 13.5-kb backbone region matching IncFII plasmid pKp_Goe_414-4Citation34 was found in pBJ20-tetA, p71221-mphA, and pW08291-tetA; a 8.3-kb one and a 25.7-kb one, a 7.4-kb one, and a 43.6-kb one matching IncFII plasmids p1220-CTXMCitation35 and pKPN1705-2 (CP022825), p16005813A,Citation36 and IncI plasmid p628-CTXMCitation37 in pW08291-tetA and pA2508-emrE, pL21-1NR, and pSC138, respectively.
Modular Differences in IncFIB-4.2 Single-Replicon Plasmids
The five IncFIB-4.2 plasmids had very similar backbones with at least two major modular differences: orf537 in pFB2.3 and pA2359-IMP was deleted due to LAR integration (Figure S2); the 43-bp tandem repeats within parC varied in copy numbers, namely 23, 21, 17, 19, and 8 for p10057-catA, p13294-1NR, pA1876-NR, pA2359-IMP, and pFB2.3, respectively.
The five LARs (Figure S2) displayed at least six major genetic differences. Firstly, ARRs were found in p10057-catA and pA2359-IMP. Secondly, ars loci were found in p10057-catA, p13294-1NR, and pFB2.3. Thirdly, sil–cop regions were present in p13294-1NR, pA1876-NR and pFB2.3. Fourthly, the IS15DI (a minor variant of IS26)-composite transposon Tn6755 was located in p13294-1NR. Fifthly, different IncFIB-4.2 plasmids had distinct profiles of metabolism gene loci: hut (histidine uptake) and ∆lac in p10057-catA; ehu (ectoine uptake), gsi and phn in p13294-1NR; ∆ehu and gsi in pA1876-NR; fec, lac and hut in pA2359-IMP; and ∆ehu in pFB2.3. Sixthly, backbone regions of IncHI1 plasmid pKP21HI1Citation38 were contained in the four plasmids except for pA1876-NR.
Diversified ARRs
The eight ARR elements ARRpA2359-IMP blaIMP-38-carrying Tn6382 (Tn21-subfamily unit transposon) as described previously,Citation39 ARRp71221-mphA, ARRpW08291-tetA, ARRpBJ20-tetA, ARRp10057-catA, ARR-1pSC138, ARR-2pSC138, and ARRpA2508-emrE varied in size from 4.1 kb to 98.7 kb.
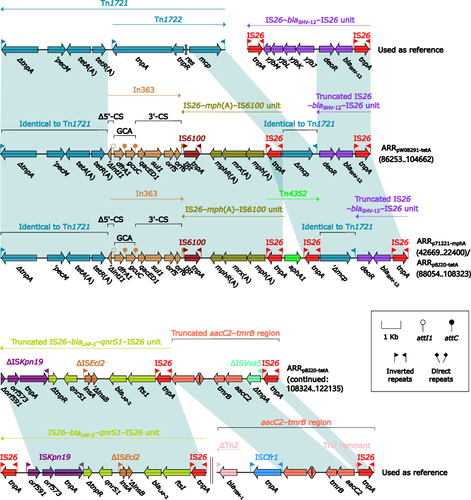
ARRpW08291-tetA, ARRp71221-mphA, and ARRpBJ20-tetA () displayed similar modular structures. ARRpW08291-tetA was derived from Tn3-family tetA(A)-carrying unit transposon Tn1721:Citation40 a tnpA (transposase)–tnpR (resolvase)–res-containing region within Tn1721 was replaced by In363Citation41 together with IS26–mph(A)–IS6100 unit;Citation42 and this modified Tn1721 further connected with a truncated IS26–blaSHV-12–IS26 unit.Citation43 ARRp71221-mphA differed from ARRpW08291-tetA by additional insertion of aphA1-carrying IS26-composite transposon Tn4352Citation44 upstream of IS26–mph(A)–IS6100 unit. ARRpBJ20-tetA contained the whole ARRp71221-mphA, with addition of a truncated IS26–blaLAP-2–qnrS–IS26 unitCitation45 and a truncated aacC2–tmrB region.Citation46
Figure 2 Organization of ARRp71221-mphA, ARRpW08291-tetA and ARRpBJ20-tetA, and comparison to related regions.
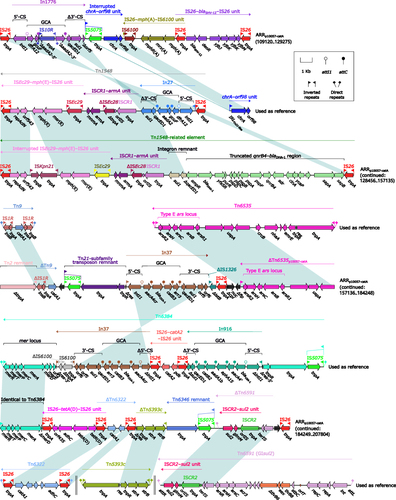
ARRp10057-catA () contained at least 12 antibiotic resistance loci: In1776, an interrupted chrA–orf98 unit, IS26–mph(A)–IS6100 unit, IS26–blaSHV-12–IS26 unit, a Tn1548-related element, a truncated IS1R-composite transposon Tn9 carrying catA1,Citation47 In37,Citation48 a truncated Tn21-subfamily unit transposon Tn6535,Citation49 IS26–tetA(D)–IS26 unit,Citation42 a truncated IS26-composite transposon Tn6322 carrying catA1,Citation50 a truncated Tn163-subfamily unit transposon Tn5393c carrying strAB,Citation51 a truncated integrative and mobilizable element Tn6591 carrying sul2.Citation52 The concise class 1 integron In1776 carried the gene cassette array (GCA) dfrA12–aadA2:IS10R–gcuF, which highly resembled that of In27.Citation53 Tn1548,Citation54 an IS26-composite transposon lack of direct repeats (DRs; target site duplication signals for transposition), contained three resistance loci: ISEc29–mph(E)–IS26 unit, ISCR1–armA unit, and In27. The Tn1548-related element had two major modular differences relative to Tn1548: ISEc29–mph(E)–IS26 unit was interrupted by insertion of ISKpn21; and a qnrB4–blaDHA-1 region was present instead of In27.
Figure 3 Organization of ARRp10057-catA, and comparison to related regions.
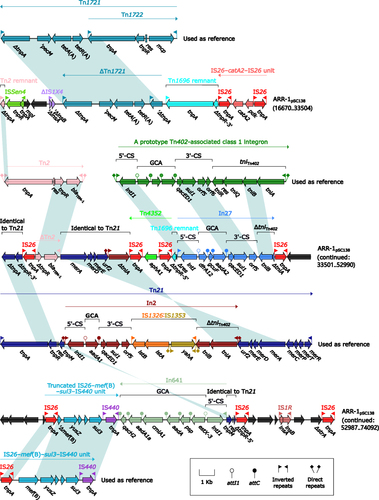
ARR-1pSC138 () harbored at least seven antibiotic resistance loci: ∆Tn1721, IS26–catA2–IS26 unit,Citation50 a truncated Tn3-subfamily unit transposon Tn2 carrying blaTEM-1,Citation55 Tn4352, In27, a truncated IS26–mef(B)–sul3–IS440 unit,Citation56 and In641.Citation57 ARR-2pSC138 (Figure S3) and ARRpA2508-emrE (Figure S4) manifested as Tn6092Citation57 (a blaCMY-2-carrying ISEcp1-based transposition unit that was another derivative of Tn6538a/b/cCitation58), and a truncated Tn21-subfamily unit transposon Tn6535 carrying emrE,Citation39 respectively.
Figure 4 Organization of ARR-1pSC138, and comparison to related regions.
Other Key Accessory Regions
Eight plasmids harbored five types of ars loci (type A to E; Figure S4): incomplete type A in ∆Tn6736 of pBJ20-tetA, p71221-mphA and pW08291-tetA, type B in pL21-1NR, type C in p13294-1NR and pFB2.3, type D in pA2508-emrE and pFB2.3, and type E in two distinct ∆Tn6535 of pA2508-emrE and p10057-catA, respectively. ∆Tn6736 and ∆Tn6535 were the truncated versions of Tn3-family prototype unit transposons Tn6736 and Tn6535,Citation39 respectively. Different types of ars loci exhibited considerable diversity in gene organization and nucleotide and amino acid sequences.
The prototype sil–cop region as initially characterized in plasmid R478Citation59 was present in three IncFIB-4 plasmids, while those in other five IncFIB-4 plasmids underwent insertion, deletion, duplication, and inversion (), especially including insertion of LARp71221-mphA, Tn6831pFB2.3 (an IS1X2-composite transposon related to cytosine metabolism), and Tn6732bpL21-1NR (a cryptic IS1F-composite transposon genetically closed related to Tn6732a).
Figure 5 Organization of sil–cop regions, and comparison to related regions.
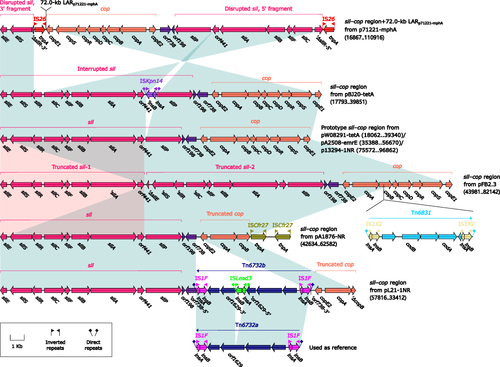
Tn6755p13294-1NR (Figure S5) was a 138.5-kb IS15DI-composite transposon and contained mainly phn, ars, sil–cop and two novel Tn3-family unit transposons Tn6756b (cryptic) and Tn6857 (glucose metabolism).
Transferability and Bacterial Antimicrobial Susceptibility
p71221-mphA or p10057-catA as the IncFIB-4.1 or IncFIB-4.1 representative plasmid could be transferred from its wild-type isolate into TOP10 through electroporation, generating E. coli electroporant TOP10/p71221-mphA or TOP10/p10057-catA, respectively. As expected, these two electroporants were resistant to azithromycin and chloramphenicol with very high minimum inhibitory concentration values ≥256 and ≥32, resp ectively. Repeated attempts to plasmid conjugal transfer failed, being consistent with the fact that these plasmids lacked conjugal transfer genes.
Novel MGEs Identified
There were totally three newly identified MGEs: integron In1776, composite transposon Tn6755, and unit transposon Tn6857. Additional seven MGEs (composite transposons Tn6732b and Tn6831, unit transposons Tn6736 and Tn6756b, and IS elements ISKqu3, ISLead2 and ISLead3) were newly designated on the basis of standard MGE nomenclatures, but they had previously determined sequences.
Concluding Remarks
In summary, a repB sequence-based typing and nomenclature scheme is proposed to identify different types or subtypes of IncFIB replicon, and a detailed comparison of the modular structures of IncFIB-4.1/4.2 single-replicon plasmids is performed to understand the roles of gene loss/acquisition in diversification of IncFIB plasmids. The IncFIB-4.1/4.2 single-replicon plasmids have rather small backbones, which undergo massive deletion events and thus display considerable modular diversification. The IncFIB-4.1/4.2 minimum backbone includes only three key backbone markers repB together with its iterons, parABC, and stbD, related to plasmid replication and maintenance. Each plasmid contains one LAR inserted into the backbone, and these LARs have significantly distinct profiles of MGEs and resistance/metabolism gene loci, thus displaying separate assembly and evolution histories. Seven of these 11 LARs contain one or two ARRs, and thus IncFIB-4.1/4.2 plasmids are important vectors of resistance genes, contributing to resistance to not only antibiotics (β-lactams including carbapenems, quinolones, aminoglycosides, tetracyclines, and phenicols) but also heavy metals (mercuric ions, copper, silver ions, and arsenic). Acquisition and accumulation of accessory regions containing resistance/metabolism markers would increase adaptability of IncFIB-4.1/4.2 plasmids-carrying bacteria under complex environments. Ten of these 11 LARs acquire one or two regions responsible for plasmid maintenance, which would facilitate stable replication of these IncFIB single-replicon plasmids at steady-state copy numbers. Data presented here provides a deeper insight into diversity and evolution of IncFIB replicon and IncFIB-4.1/4.2 single-replicon plasmids.
Data Sharing Statement
The datasets generated for this study can be found in the complete nucleotide sequences of plasmids pBJ20-tetA, p71221-mphA, pW08291-tetA, pA2508-emrE, pL21-1NR, p10057-catA, p13294-1NR, pA1876-NR, and pA2359-IMP were submitted to GenBank under accession numbers MN310373, MN310374, MN310376, MN310379, MN423365, MN423364, MT570100, MT549899, and MN423363, respectively.
Ethics Statement
This study uses the clinical bacterial isolates obtained from the Chinese public hospitals as listed in Table S1. The local legislation did not require the study to be reviewed or approved by an ethics committee, because the bacterial isolates involved in this study was part of the routine hospital Laboratory procedures. The research involving biohazards and all related procedures were approved by the Biosafety Committee of the Beijing Institute of Microbiology and Epidemiology.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
Acknowledgments
All experiments and data analyses were done in Dr. Dongsheng Zhou’s laboratory.
Additional information
Funding
References
- Lederberg J, Tatum EL. Gene recombination in Escherichia coli. Nature. 1946;158(4016):558. doi:10.1038/158558a0
- Yang QE, Sun J, Li L, et al. IncF plasmid diversity in multi-drug resistant Escherichia coli strains from animals in China. Front Microbiol. 2015;6:964. doi:10.3389/fmicb.2015.00964
- Tang Y, Fu P, Zhou Y, et al. Absence of the type I-E CRISPR-Cas system in Klebsiella pneumoniae clonal complex 258 is associated with dissemination of IncF epidemic resistance plasmids in this clonal complex. J Antimicrob Chemother. 2020;75(4):890–895. doi:10.1093/jac/dkz538
- Carattoli A, Villa L, Pezzella C, Bordi E, Visca P. Expanding drug resistance through integron acquisition by IncFI plasmids of Salmonella enterica Typhimurium. Emerg Infect Dis. 2001;7(3):444–447. doi:10.3201/eid0703.017314
- Chen L, Chavda KD, Melano RG, et al. Complete sequence of a blaKPC-2-harboring IncFIIK1 plasmid from a Klebsiella pneumoniae sequence type 258 strain. Antimicrob Agents Chemother. 2013;57(3):1542–1545. doi:10.1128/AAC.02332-12
- Du XD, Li DX, Hu GZ, et al. Tn1548-associated armA is co-located with qnrB2, aac(6’)-Ib-cr and blaCTX-M-3 on an IncFII plasmid in a Salmonella enterica subsp. enterica serovar Paratyphi B strain isolated from chickens in China. J Antimicrob Chemother. 2012;67(1):246–248. doi:10.1093/jac/dkr407
- Ho PL, Chan J, Lo WU, Law PY, Chow KH. Plasmid-mediated fosfomycin resistance in Escherichia coli isolated from pig. Vet Microbiol. 2013;162(2–4):964–967. doi:10.1016/j.vetmic.2012.09.023
- Lam MMC, Wyres KL, Duchêne S, et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun. 2018;9(1):2703. doi:10.1038/s41467-018-05114-7
- García-Fernández A, Villa L, Carta C, et al. Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob Agents Chemother. 2012;56(4):2143–2145. doi:10.1128/AAC.05308-11
- Villa L, Feudi C, Fortini D, et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb Genom. 2017;3(4):e000110. doi:10.1099/mgen.0.000110
- Villa L, García-Fernández A, Fortini D, Carattoli A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother. 2010;65(12):2518–2529. doi:10.1093/jac/dkq347
- Saadi S, Maas WK, Hill DF, Bergquist PL. Nucleotide sequence analysis of RepFIC, a basic replicon present in IncFI plasmids P307 and F, and its relation to the RepA replicon of IncFII plasmids. J Bacteriol. 1987;169(5):1836–1846. doi:10.1128/jb.169.5.1836-1846.1987
- Chattoraj DK. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol Microbiol. 2000;37(3):467–476. doi:10.1046/j.1365-2958.2000.01986.x
- Li JJ, Spychala CN, Hu F, Sheng JF, Doi Y. Complete nucleotide sequences of blaCTX-M-harboring IncF plasmids from community-associated Escherichia coli strains in the United States. Antimicrob Agents Chemother. 2015;59(6):3002–3007. doi:10.1128/AAC.04772-14
- Liu P, Li P, Jiang X, et al. Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J Bacteriol. 2012;194(7):1841–1842. doi:10.1128/JB.00043-12
- Qu D, Shen Y, Hu L, et al. Comparative analysis of KPC-2-encoding chimera plasmids with multi-replicon IncR:IncpA1763-KPC: incN1or IncFIIpHN7A8:IncpA1763-KPC:IncN1. Infect Drug Resist. 2019;12:285–296. doi:10.2147/IDR.S189168
- Osborn AM, da Silva Tatley FM, Steyn LM, Pickup RW, Saunders JR. Mosaic plasmids and mosaic replicons: evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology. 2000;146(Pt 9):2267–2275. doi:10.1099/00221287-146-9-2267
- Oshima K, Toh H, Ogura Y, et al. Complete genome sequence and comparative analysis of the wild-type commensal Escherichia coli strain SE11 isolated from a healthy adult. DNA Res. 2008;15(6):375–386. doi:10.1093/dnares/dsn026
- Nazir A, Zhao Y, Li M, et al. Structural Genomics of repA, repB1-carrying IncFIB family pA1705-qnrS, p911021-tetA, and p1642-tetA, multidrug-resistant plasmids from Klebsiella pneumoniae. Infect Drug Resist. 2020;13:1889–1903. doi:10.2147/IDR.S228704
- Fu J, Zhang J, Yang L, et al. Precision methylome and in vivo methylation kinetics characterization of Klebsiella pneumoniae. Genomics Proteomics Bioinformatics. 2021. doi:10.1016/j.gpb.2021.04.002
- Hackl T, Hedrich R, Schultz J, Förster F. proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics. 2014;30(21):3004–3011. doi:10.1093/bioinformatics/btu392
- Brettin T, Davis JJ, Disz T, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5:8365. doi:10.1038/srep08365
- Boratyn GM, Camacho C, Cooper PS, et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013;41:W29–33. doi:10.1093/nar/gkt282
- O’Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi:10.1093/nar/gkv1189
- Carattoli A, Zankari E, García-Fernández A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–3903. doi:10.1128/AAC.02412-14
- Jia B, Raphenya AR, Alcock B, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45(D1):D566–D573. doi:10.1093/nar/gkw1004
- Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. doi:10.1093/jac/dks261
- Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi:10.1093/nar/gkj014
- Moura A, Soares M, Pereira C, Leitão N, Henriques I, Correia A. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics. 2009;25(8):1096–1098. doi:10.1093/bioinformatics/btp105
- Roberts AP, Chandler M, Courvalin P, et al. Revised nomenclature for transposable genetic elements. Plasmid. 2008;60(3):167–173. doi:10.1016/j.plasmid.2008.08.001
- Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27(1):135–145. doi:10.1002/pro.3290
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi:10.1093/molbev/msy096
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
- Wang S, Dai E, Jiang X, et al. Characterization of the plasmid of incompatibility groups IncFIIpKF727591 and IncpKPHS1 from Enterobacteriaceae species. Infect Drug Resist. 2019;12:2789–2797. doi:10.2147/IDR.S212321
- Zhang D, Yin Z, Zhao Y, et al. p1220-CTXM, a pKP048-related IncFIIK plasmid carrying blaCTX-M-14 and qnrB4. Future Microbiol. 2017;12:1035–1043. doi:10.2217/fmb-2017-0026
- Yin Z, Hu L, Cheng Q, et al. First report of coexistence of three different MDR plasmids, and that of occurrence of IMP-encoding plasmid in Leclercia adecarboxylata. Front Microbiol. 2019;10:2468. doi:10.3389/fmicb.2019.02468
- Wang L, Fang H, Feng J, et al. Complete sequences of KPC-2-encoding plasmid p628-KPC and CTX-M-55-encoding p628-CTXM coexisted in Klebsiella pneumoniae. Front Microbiol. 2015;6:838. doi:10.3389/fmicb.2015.00838
- Li L, Yu T, Ma Y, et al. The genetic structures of an extensively drug resistant (XDR) Klebsiella pneumoniae and its plasmids. Front Cell Infect Microbiol. 2018;8:446. doi:10.3389/fcimb.2018.00446
- Liang Q, Jiang X, Hu L, et al. Sequencing and genomic diversity analysis of IncHI5 plasmids. Front Microbiol. 2018;9:3318. doi:10.3389/fmicb.2018.03318
- Allmeier H, Cresnar B, Greck M, Schmitt R. Complete nucleotide sequence of Tn1721: gene organization and a novel gene product with features of a chemotaxis protein. Gene. 1992;111(1):11–20. doi:10.1016/0378-1119(92)90597-I
- Levings RS, Djordjevic SP, Hall RM. SGI2, a relative of Salmonella genomic island SGI1 with an independent origin. Antimicrob Agents Chemother. 2008;52(7):2529–2537. doi:10.1128/AAC.00189-08
- Liang Q, Yin Z, Zhao Y, et al. Sequencing and comparative genomics analysis of the IncHI2 plasmids pT5282-mphA and p112298-catA and the IncHI5 plasmid pYNKP001-dfrA. Int J Antimicrob Agents. 2017;49(6):709–718. doi:10.1016/j.ijantimicag.2017.01.021
- Ford PJ, Avison MB. Evolutionary mapping of the SHV beta-lactamase and evidence for two separate IS26-dependent blaSHV mobilization events from the Klebsiella pneumoniae chromosome. J Antimicrob Chemother. 2004;54(1):69–75. doi:10.1093/jac/dkh251
- Wrighton CJ, Strike P. A pathway for the evolution of the plasmid NTP16 involving the novel kanamycin resistance transposon Tn4352. Plasmid. 1987;17(1):37–45. doi:10.1016/0147-619X(87)90006-0
- Le V, Nhu NTK, Cerdeno-Tarraga A, et al. Genetic characterization of three qnrS1-harbouring multidrug-resistance plasmids and qnrS1-containing transposons circulating in Ho Chi Minh City, Vietnam. J Med Microbiol. 2015;64(8):869–878. doi:10.1099/jmm.0.000100
- Partridge SR, Ginn AN, Paulsen IT, Iredell JR. pEl1573 carrying blaIMP-4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob Agents Chemother. 2012;56(11):6029–6032. doi:10.1128/AAC.01189-12
- Alton NK, Vapnek D. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature. 1979;282(5741):864–869. doi:10.1038/282864a0
- Wang M, Tran JH, Jacoby GA, Zhang Y, Wang F, Hooper DC. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob Agents Chemother. 2003;47(7):2242–2248. doi:10.1128/AAC.47.7.2242-2248.2003
- Tan JY, Yin WF, Chan KG. Gene clusters of Hafnia alvei strain FB1 important in survival and pathogenesis: a draft genome perspective. Gut Pathog. 2014;6:29. doi:10.1186/1757-4749-6-29
- Shi L, Liang Q, Zhan Z, et al. Co-occurrence of 3 different resistance plasmids in a multi-drug resistant Cronobacter sakazakii isolate causing neonatal infections. Virulence. 2018;9(1):110–120. doi:10.1080/21505594.2017.1356537
- L’Abée-Lund TM, Functional SH. Tn5393-like transposon in the R plasmid pRAS2 from the fish pathogen Aeromonas salmonicida subspecies salmonicida isolated in Norway. Appl Environ Microbiol. 2000;66(12):5533–5535. doi:10.1128/AEM.66.12.5533-5535.2000
- Harmer CJ, Hamidian M, Hall RM. pIP40a, a type 1 IncC plasmid from 1969 carries the integrative element GIsul2 and a novel class II mercury resistance transposon. Plasmid. 2017;92:17–25. doi:10.1016/j.plasmid.2017.05.004
- Partridge SR, Paulsen IT, Iredell JR. pJIE137 carrying blaCTX-M-62 is closely related to p271A carrying blaNDM-1. Antimicrob Agents Chemother. 2012;56(4):2166–2168. doi:10.1128/AAC.05796-11
- Galimand M, Sabtcheva S, Courvalin P, Lambert T. Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob Agents Chemother. 2005;49(7):2949–2953. doi:10.1128/AAC.49.7.2949-2953.2005
- Partridge SR. What’s in a name? ISSwi1 corresponds to transposons related to Tn2 and Tn3. mBio. 2015;6(5):e01344–e01315. doi:10.1128/mBio.01344-15
- Liu J, Keelan P, Bennett PM, Enne VI. Characterization of a novel macrolide efflux gene, mef(B), found linked to sul3 in porcine Escherichia coli. J Antimicrob Chemother. 2009;63(3):423–426. doi:10.1093/jac/dkn523
- Ye J, Su LH, Chen CL, et al. Analysis of pSC138, the multidrug resistance plasmid of Salmonella enterica serotype Choleraesuis SC-B67. Plasmid. 2011;65(2):132–140. doi:10.1016/j.plasmid.2010.11.007
- Cheng Q, Jiang X, Xu Y, et al. Type 1, 2, and 1/2-hybrid IncC plasmids from China. Front Microbiol. 2019;10:2508. doi:10.3389/fmicb.2019.02508
- Gilmour MW, Thomson NR, Sanders M, Parkhill J, Taylor DE. The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid. 2004;52(3):182–202. doi:10.1016/j.plasmid.2004.06.006
- Partridge SR, Brown HJ, Stokes HW, Hall RM. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob Agents Chemother. 2001;45(4):1263–1270. doi:10.1128/AAC.45.4.1263-1270.2001
