Abstract
Introduction
In recent decades, the rate of infection with dengue virus (DENV) has risen significantly, now affecting nearly 400 million individuals annually. Dengue fever among humans is caused via specific mosquito vectors bites. Sporadic cases have been reported in Egypt. The goal of this study was to identify the serotype of the DENV outbreak in both human and mosquito vector along the coast of the Red Sea, Upper Egypt, in 2017. Identification of the serotype of the virus may help identify its source and assist in applying epidemiological and control measures.
Materials and Methods
The current study was carried out in El Quseir City, Red Sea Governorate, Upper Egypt, on 144 patients complaining of symptoms indicative of dengue fever at the time of the 2017 Egypt outbreak. Human blood samples and the mosquito reservoirs were identified as having dengue virus infection serologically and molecularly.
Results
Overall, 97 (67.4%) patients were positive for dengue virus IgM antibodies. Molecular examination of the human samples and pools of mosquitoes revealed that DENV-2 virus was the serotype responsible for the outbreak. Only one pool of female mosquitoes containing Aedes aegypti was infected with dengue fever virus (DENV-2).
Conclusion
This was the first serotyping of the DENV responsible for dengue virus outbreak in Egypt in 2017. Determining the serotype of dengue virus can help to avoid and monitor outbreaks. The serotype identified in this study was DENV-2, while DENV-1 was the serotype found in the previous outbreak in 2015 in the province of Assiut. This study thus raises concerns that a new dengue serotype could have been introduced into Egypt. It is necessary for a comprehensive risk assessment to be carried out in the country, including an entomological survey, to assess the presence and potential geographical expansion of mosquito vectors there.
Introduction
Dengue virus (DENV) that belongs to the family Flaviviridae, genus Flavivirus, is the causative agent for human dengue fever. This is an arboviral disease spread via the biting of female mosquito vectors (Aedesa egypti and Ae. Albopictus).Citation1,Citation2 The infection rate has grown globally especially during rainy seasons in urban and suburban regions in Asian countries, and affecting approximately 400 million individuals annually.Citation3–5 The World Health Organization (WHO) statistics reported that about 50% of the world’s population is at risk of infection.Citation6
Since the nineteenth century, dengue cases have been reported in several African countries, including Egypt.Citation7 Years later, more outbreaks have been documented in fifteen countries, especially in East Africa. The main isolated DENV serotype in African epidemics was DENV-2.Citation7
DENV has a single-stranded RNA genome and includes four serotypes (DENV-1, DENV-2, DENV-3 and DENV-4) that causes a self-limiting febrile illness called dengue fever and lasting for few days.Citation8 Some cases can complicate to life-threatening conditions characterized by hemorrhage and thrombocytopenia (dengue hemorrhagic fever), or excessive plasma leakage (dengue shock syndrome).Citation9 The rate of mortality varies across reported cases, with a mean of 5% in general population infected with DENV.Citation10
DENV rapid evolution is associated with the “error-prone RNA-dependent RNA polymerase”, which lead to the formation of several genotypes in each serotype.Citation8,Citation11 This evolution phenomena has been associated with epidemic activity, and sometimes with the intensity of the infection.Citation3,Citation12,Citation13 Other factors, such as the genetic constitution of DENV, the genetic make-up of the patient, were correlated with life-threatening conditions, in addition to previous infection with a different serotype.Citation14 The spread of the mosquito vector combined with increased urbanization can lead to hyperendemicity (multiple virus serotypes co-circulating). Despite challenges facing the development of dengue vaccine including the different serotypes and the cross reactivity with other Flavivirus, several attempts revealed promising results, in addition to the current vector control strategies.Citation15–17
The aim of our study was to identify the serotypes of DENV in the outbreak that took place in Red Sea Governorate, Upper Egypt, in 2017 in both human and mosquitoes, in addition to identifying the mosquito vector involved during the outbreak. The present study is the first to identify DENV serotypes in the 2017 Egyptian outbreak in either humans or mosquitoes. This will identify the source of the virus and aid the implementation of epidemiological and control strategies.
Materials and Methods
Study Area
This study was carried out in El Quseir City (145 km south of Hurghada), Red Sea Governorate, Upper Egypt, in the period between the 1st of October and 30th of December 2017 (at the time of the 2017 dengue fever outbreak in Egypt) ().
Figure 1 Map of Egypt displaying the location of the Red Sea Governorate and its cities. Site of the study is Quseir City adapted from https://freesvg.org/1546672123.Citation18
Collection and Examination of Human Samples
We included 144 patients who attended Safaga General Hospital, Red Sea Governorate, Egypt. All participants complaining of febrile illness with at least two of the following manifestations: myalgia, arthralgia, headache, and skin rashes were included. All patients underwent a comprehensive interview about their medical history (age, sex, myalgia, arthralgia, rashes, and previous similar manifestations) and a clinical examination (temperature, blood pressure, and bleeding tendency). Blood samples were collected, in ethylenediaminetetraacetic acid (EDTA) containing tubes, after which the plasma was separated by centrifugation and divided into two parts: one part was used for detecting anti-dengue virus Immunoglobulin M (IgM) and the other part was kept at −20℃ for further molecular examination.
Diagnosis of Dengue Infection
Dengue virus IgM was estimated using ELISA kit in accordance with the manufacturer’s instructions (Abcam, UK).
Molecular Analysis
RNA Extraction
Plasma samples of IgM-positive cases were subjected to RNA extraction using QIAamp Viral RNA Extraction Kit (Cat# 219610), in accordance with the instructions of the manufacturer. Briefly, each sample was vortexed properly with 5 volumes of lysis reagent, then the homogenate was incubated at (15–25℃) for 5 min and followed by addition of miRNeasy serum/plasma Spike-In control and chloroform. After vigorous shaking and incubation at room temperature for 2–3 min, the aliquot was centrifuged at 8000 rpm to separate the sample into 3 layers. The top colorless aqueous layer (contains RNA) was mixed well in another tube containing 1.5 volumes of absolute ethyl alcohol. Using an RNeasy Mini-Elute spin column, the mixture was centrifuged at 8000 rpm and the supernatant was discarded. This step was repeated for 3 times. To dry the membrane, 500 µL of 80% ethanol was added to the spin column, then centrifuged at high speed for 5 min. To elute the RNA, 14 µL RNase-free distilled water was added to the spin column membrane, then centrifuged at high speed for 1 min. ND-1000 UV-Vis Spectrophotometer (NanoDrop®) was used to estimate the concentration of the RNA (ng/μL) after extraction.
First Step Amplification: Reverse Transcription Polymerase Chain Reaction (RT-PCR) of Dengue Virus RNA
Using the SuperScript III First-Strand Synthesis System (Invitrogen, USA), dengue virus RNA was converted into a cDNA, which then was used for virus amplification using D1 consensus forward primer and D2 reverse primer. The PCR product was then used as the template for the second step to detect the different DENV serotypes.
Second Step Amplification: Multiplex PCR with Serotype-Specific Primers
This amplification was performed to detect the four serotypes. PCR products from the first step of the RT-PCR were used as templates. D1 primer was used as a forward primer (conserved in all dengue serotypes). The specific oligonucleotides (TS1, TS2, TS3, and TS4) for each serotype (DEN-1, DEN-2, DEN-3, and DEN respectively) as reverse primersCitation19 ().
Table 1 The Five Oligonucleotide Primers Used in Multiplex PCR for Identification of the DENV Serotype in the Present Study
The total reaction volume was 25 µL, consisting of the following: 5 µL of the extracted RNA, 5 µL of 5× Qiagen RT-PCR buffer, 1 µL of dNTP mix, 1 µL of Qiagen RT-PCR enzyme mix (reverse transcriptase + DNA polymerase), 1 µL of each primer (D1, TS1, TS2, TS3, and TS4), and 8 µL of RNase-free distilled water. The thermal cycling (peqSTAR; VWR International, UK) first step involved activation of reverse transcriptase enzyme to convert viral RNA to DNA, in which the temperature was set to 50℃ for 30 min. The second step involved DNA amplification, which featured hot-start at 95℃ for 15 min for activation of DNA polymerase enzyme, followed by 40 cycles of initial denaturation (1 min at 94℃), annealing (1 min at 55℃), and primer extension (1 min at 72℃), and final extension at 72℃ for 10 min.
The PCR products and a 100 bp DNA ladder were run on 2% agarose gel containing ethidium bromide, then visualized under UV. The imaging was carried out with a Gel Documentation System (Cleaver Scientific, UK). The expected molecular weights for the four serotypes DENV-1-4 were 482 bp, 119 bp, 290 bp, and 389 bp, respectively ().
Table 2 The Oligonucleotide Primer Set for Each Virus Serotype and Expected Product Size
Collection and Examination of Mosquitoes
After identification of positive dengue virus-related IgM antibody human cases, a total of 193 mosquitoes were collected using CDC light traps between October and December 2017. Twenty collection sites were distributed throughout Quseir City, where clinical cases were reported. Collected specimens were labeled with location and date of capture, stored on dry ice then sent to the department of Medical Parasitology, Faculty of Medicine, South Valley University, and stored at −80℃ until further processing.Citation20–22 Mosquitoes were identified according to.Citation23 Identified mosquitoes were then pooled according to species and sex into 10 pools, each contained 19–20 insects.
Insects in each pool were macerated in 100 μL of phosphate-buffered saline before RNA extraction in a 1:1 phenol and chloroform mixture. For each sample, 5 μL of acid-washed size-selected silica particles were mixed, then after five minutes centrifuged, and the formed pellet was washed twice (10 mM Tris pH 7.4, 50% ethanol, 50 mM NaCl, 1 mM EDTA). The pellet was resuspended in 10 μL of RNase-free distilled water, incubated for 5 min at 55℃, then centrifuged. The resulted supernatant eluate was transferred to another tube. Immediately prior amplification, the washed pellet was resuspended in 5 μL of RNase-free distilled water.Citation24 Molecular examination of mosquito samples was performed using the same protocol for the human samples.
Statistical Analysis
All obtained data were statistically analyzed by IBM SPSS, version 22. Quantitative data were expressed as mean ± standard deviation. Qualitative data were expressed as number and percentage.
Results
The mean age of the included 144 patients was 41.4 ± 13.7. Ninety-seven (67.4%) were male. All patients presented fever. Other clinical manifestations included headache, arthralgia, myalgia, and rash (bleeding tendency). The epidemiological data and clinical presentation of the participants are shown in .
Table 3 Epidemiological Data and Clinical Manifestations of Patients Participating in the Present Study
From the serological examination of human plasma, 97 (67.4%) patients were positive for DENV IgM (). Positive samples were subjected to further molecular examination using RT Multiplex PCR analysis. After the RNA extraction, we measured the RNA concentration in the samples using Nanodrop. RNA concentration in samples ranged from 4 to 151.4 ng/µL - with 260/280 ratio from 1.7 to 3.3. Samples with low concentration of RNA gave negative results for multiplex PCR; however, samples with RNA concentration form 21–114.8 ng/µL were positive using multiplex PCR (63 samples) ().
Table 4 Results of ELISA/IgM Examination of Patients Samples Layered by Age and Gender
Table 5 Results of Multiplex PCR Examination of ELISA/IgM Positive Samples Layered by Age and Gender
Molecular analysis of the human samples showed that the virus serotype that was responsible for the outbreak was DENV-2. In all examined samples, the bands resulting from amplification using the five oligonucleotide primers were at 119 bp, indicating infection with DENV-2 virus ().
Figure 2 Agarose gel electrophoretic analysis of DENG samples amplified by One-Step Multiplex RT-PCR. (A) Human samples; amplified products of clinical samples, positive samples showing 119 bp (DENV-2). (B) Amplified products of Adult female Aedes aegypti, positive samples showing 119 bp (DENV-2). DNA ladder 100 bp.
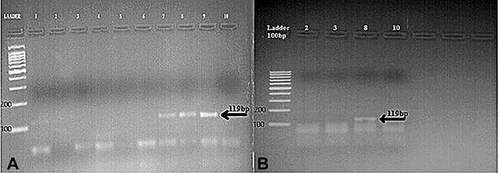
As shown in , a total of 193 mosquitoes were captured. Microscopic examination of mosquitoes revealed four species, namely; Ae. aegypti, Culex pipiens, Anopheles sergentii, and An. multicolor. Only one pool of adult female Ae. Aegebti was identified, and then used for Molecular analysis. Bands resulting from amplification were at 119 bp, indicating infection with DENV-2 virus ().
Table 6 Species of Mosquitoes Tested for Arboviruses, Collected Between October 2017 and December 2017, Quseir City, Red Sea Governorate, Egypt
Discussion
The factors that contribute to the exacerbation of the problem of dengue fever are the random unplanned overcrowding of the population, climate change, and widespread population movements. The unprecedented increase in flights has led to the spread of arboviruses and related vectors into new areas and to disease-naive human populations. Dengue fever is now considered among the main causes for post-travel fever.Citation25
Periodic outbreaks of dengue fever have occurred in Egypt with an increasing number of cases of increasing severity. The first outbreak of dengue fever in Egypt was reported in 1799 in Cairo and Alexandria governorates. This was followed by an outbreak reported in Port Said in 1871, an outbreak reported in Cairo in 1880, an outbreak reported in the Nile Delta in 1889, an outbreak in Port Said and the Suez Canal in 1906, an outbreak throughout Egypt (including Upper Egypt) in 1927, an outbreak in Egypt (area unknown) in 1937, travel-associated cases reported in Italy in two travelers returning from a Red Sea resort in 2010, and an outbreak in Assiut Governorate in 2015.Citation26
The current study was conducted during a recent outbreak of dengue fever in Red Sea Governorate, Upper Egypt, in 2017. A total of 144 patients with subjective symptoms participated in the present study. The mean age of our patients was 41.4 ± 13.74, which is in accordance with previous studies.Citation27,Citation28 As reported previously, infection among males was is higher than females.Citation28–30 In our study, all patients complained of headache and fever, while more around 90% presented arthralgia and myalgia. In relation to more virulent genotype, previous workers noticed higher frequency of hemorrhagic manifestations including epistaxis, gum bleeding, melena, and even hematemesis and skin rashes.Citation30,Citation31
Positive samples were subjected to Multiplex RT PCR. DENV-2 serotype was the reported serotype in both humans and mosquitoes. Our study is the first to identify the DENV serotype causative of the Egyptian outbreak in 2017. Dengue fever outbreaks and epidemics are not uncommon in Africa, with recent outbreaks occurring predominantly in the east of the continent.Citation7 Egypt is bordered to the east by the Red Sea with Saudi Arabia and to the south by Sudan, where these countries were declared endemic for dengue fever and frequently suffer from dengue outbreaks. The predominant three serotypes of the virus (DENV-1, DENV-2, and DENV-3) were documented in several Middle East countries including Saudi Arabia,Citation31 particularly Jeddah and Makkah. It was reported that DENV-2 infection was more common than DENV-1 or DENV-3 infection in Jeddah and Makkah.Citation32,Citation33 Recently several outbreaks and sporadic cases has spread in many areas in Sudan.Citation34–36 DENV can also be introduced by travelers from endemic regions.Citation37–39 DENV-1, DENV-2, and DENV-3 were confirmed to be endemic in some parts of Sudan.Citation34,Citation40
In terms of the mosquito vectors, dengue virus was identified only in female Ae. aegypti. The same virus serotype that was identified in human samples was identified in the mosquito population in our study. Earlier before 1940, Ae. aegypti species was correlated with all dengue outbreaks in Egypt, but the use of insecticides such as DDT led to a strong decrease and disappearance of mosquito population, however, studies reviewed the re-emergence of Ae. aegypti, in Egypt.Citation41,Citation42 In 2018, Abozeid et al reported the first detection of Ae. aegypti from the dengue outbreak in Red Sea Governorate,Citation41 which could be exported on ships from Saudi Arabia or via ground traffic from Sudan.
The serotype reported in the present study is DENV-2, while the serotype identified in the previous outbreak in 2015 in Assiut Governorate was DENV-1.Citation26 This raises concerns about the introduction of a new dengue serotype into Egypt. Considering the public health risk of the disease becoming endemic in Egypt, it is important that a detailed risk assessment of the situation is performed in the country, including an entomological survey to determine the existence and possible geographic expansion of mosquito vectors in the country.
We believe that many outbreaks in Egypt have not been well characterized. This could be due to the weak surveillance infrastructure and unrecognition of dengue fever. On the other hand, the reemergence of Ae. aegypti vector in Egypt may increase the risk of transmission of many arboviruses, including dengue fever. Vector control strategic approach should be considered and including effective community engagement in accordance with the WHO new strategy (2017–2030) to strengthen vector control worldwide.
Limitations of the Present Study
The only limitation of this study is the number of samples used for Multiplex PCR. ELISA/IgM positive samples only were used for DENV serotyping, in addition to the low concentration of RNA in clinical samples which led to serotyping of 63 out of 144 suspected cases. Although, a small number of samples were used, we revealed a new DENV serotype (DENV-2), which raises concerns about the introduction of a new dengue serotype into Egypt and necessitates more studies to be conducted on serotyping of different dengue serotypes in Egypt.
Ethical Statement
All procedures performed in the present study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the Declaration of Helsinki. This research received ethical approval from the institutional review board of the Faculty of Medicine, South Valley University, Egypt (SVU-MED-MIC007-3-157). Informed consent was obtained from all adult patients and the guardians of children below 18 years participating in the study.
Disclosure
The authors declare that they have no conflicts of interest.
Acknowledgment
This research was supported by Princess Nourah Bint Abdulrahman University Project number (PNURSP2022R39), Riyadh, Saudi Arabia.
References
- Khan J, Ghaffar A, Khan SA. The changing epidemiological pattern of dengue in Swat, Khyber Pakhtunkhwa. PLoS One. 2018;13(4):1–14. doi:10.1371/journal.pone.0195706
- Ferreira-de-lima VH, Lima-Camara TN. Natural vertical transmission of dengue virus in Aedes aegypti and Aedes albopictus: a systematic review. Parasit Vectors. 2018;11(1):1–8. doi:10.1186/s13071-018-2643-9
- Bennett SN, Holmes EC, Chirivella M, et al. Molecular evolution of dengue 2 virus in Puerto Rico: positive selection in the viral envelope accompanies clade reintroduction. J Gen Virol. 2006;87(Pt4):885–893. doi:10.1099/vir.0.81309-0
- Bhatt S, Gething PW, Brady OJ, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi:10.1038/nature12060
- Brady OJ, Gething PW, Bhatt S, et al. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl Trop Dis. 2012;6(8):e1760. doi:10.1371/journal.pntd.0001760
- Atique S, Chan TC, Chen CC, et al. Investigating spatio-temporal distribution and diffusion patterns of the dengue outbreak in Swat, Pakistan. J Infect Public Health. 2018;11(4):550–557. doi:10.1016/j.jiph.2017.12.003
- Amarasinghe A, Kuritsk JN, Letson GW, Margolis HS. Dengue virus infection in Africa. Emerg Infect Dis. 2011;17(8):1349–1354. doi:10.3201/eid1708.101515
- Weaver SC, Vasilakis N. Molecular evolution of dengue viruses: contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2009;9(4):523–540. doi:10.1016/j.meegid.2009.02.003
- Gubler DJ. Perspectives on the prevention and control of dengue hemorrhagic fever. Gaoxiong Yi Xue Ke Xue Za Zhi. 1994;10(Suppl):S15–8.
- Martin E, Chirivella M, Co JKG, et al. Insights into the molecular evolution of dengue virus type 4 in Puerto Rico over two decades of emergence. Virus Res. 2016;213:23–31. doi:10.1016/j.virusres.2015.11.009
- Waman VP, Kolekar P, Ramtirthkar MR, Kale MM, Kulkarni-Kale U. Analysis of genotype diversity and evolution of dengue virus serotype 2 using complete genomes. PeerJ. 2016;4:e2326. doi:10.7717/peerj.2326
- Bennett SN, Holmes EC, Chirivella M, et al. Selection-driven evolution of emergent dengue virus. Mol Biol Evol. 2003;20(10):1650–1658. doi:10.1093/molbev/msg182
- Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11(3):480–496. doi:10.1128/CMR.11.3.480
- Halstead SB. Is there an inapparent dengue explosion? Lancet. 1999;353(9158):1100–1101. doi:10.1016/S0140-6736(05)76460-6
- Webster DP, Farrar J, Rowland-Jones S. Progress towards a dengue vaccine. Lancet Infect Dis. 2009;9(11):678–687. doi:10.1016/S1473-3099(09)70254-3
- Murray NEA, Quam MB, Wilder-Smith A. Epidemiology of dengue: past, present and future prospects. Clin Epidemiol. 2013;5:299–309. doi:10.2147/CLEP.S34440
- Thomas SJ, Rothman AL. Trials and tribulations on the path to developing a dengue vaccine. Vaccine. 2015;33(Suppl 4):D24–31. doi:10.1016/j.vaccine.2015.05.095
- Free*SVG [homepage on the Internet].Egypt, administrative divisions map. 2019-2020. Available from https://freesvg.org/1546672123. Accessed May 23, 2022.
- Harris E, Roberts TG, Smith L, et al. Typing of dengue viruses in clinical specimens and mosquitoes by single-tube multiplex reverse transcriptase PCR. J Clin Microbiol. 1998;36(9):2634–2639. doi:10.1128/JCM.36.9.2634-2639.1998
- Rattanarithikul R, Harbach RE, Harrison BA, Panthusiri P, Coleman RE, Richardson JH. Illustrated keys to the mosquitoes of Thailand. VI. Tribe Aedini. Southeast Asian J Trop Med Public Health. 2010;41(Suppl 1):1–225.
- Rattanarithikul R, Harrison BA, Harbach RE, Panthusiri P, Coleman RE, Panthusiri P. Illustrated keys to the mosquitoes of Thailand. IV. Anopheles. Southeast Asian J Trop Med Public Health. 2006;37(Suppl 2):1–128.
- Consoli RAGB, Oliveira de RL. Mosquitos de Importância Sanitária Do Brasil.(Main mosquitoes of sanitary importance in Brazil)Pdf. Editora Fiocruz; 1994.
- Rueda LM. Pictorial keys for the identification of mosquitoes (Diptera:Culicidae) associated with dengue virus transmission. Zootaxa. 2004;589:1–60. doi:10.11646/zootaxa.589.1.1
- Vogelstein B, Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci USA. 1979;76(2):615–619. doi:10.1073/pnas.76.2.615
- Gloria-Soria A, Armstrong PM, Powell JR, Turner PE. Infection rate of Aedes aegypti mosquitoes with dengue virus depends on the interaction between temperature and mosquito genotype. Proceedings Biol Sci. 2017;284(1864):20171506. doi:10.1098/rspb.2017.1506
- World Health Organaization. Weekly epidemiological monitor. World Heal Organ Cairo, Egypt. 2016;6(52):22765273.
- Azhar Z, Hassan MR. temporal spatial distribution of dengue and implications on control in Hulu Langat, Selangor, Malaysia. Dengue Bull. 2016;39:19–31.
- Eldigail MH, Adam GK, Babiker RA, et al. Prevalence of dengue fever virus antibodies and associated risk factors among residents of El-Gadarif state, Sudan. BMC Public Health. 2018;18(1):1–8. doi:10.1186/s12889-018-5853-3
- Banerjee A, Barik KL, Bandyopadhyay A, Paul UK. A study on the clinical features of dengue virus infected pediatric patients. Int J Contemp Pediatr. 2018;5(2):368. doi:10.18203/2349-3291.ijcp20180437
- Khan AH, Hayat AS, Masood N, Solangi NM, Shaikh TZ. Frequency and clinical presentation of dengue fever at tertiary care hospital of Hyderabad/Jamshoro. J Liaquat Univ Med Heal Sci. 2010;9(2):88–94.
- Nedjadi T, El-Kafrawy S, Sohrab SS, Desprès P, Damanhouri G, Azhar E. Tackling dengue fever: current status and challenges Positive-strand RNA viruses. Virol J. 2015;12(1):1–11. doi:10.1186/s12985-015-0444-8
- Fakeeh M, Zaki AM. Dengue in Jeddah, Saudi Arabia, 1994–2002. Dengue Bull. 2003;27(September):13–18.
- Khan NA, Azhar EI, El-Fiky S, et al. Clinical profile and outcome of hospitalized patients during first outbreak of dengue in Makkah, Saudi Arabia. Acta Trop. 2008;105(1):39–44. doi:10.1016/j.actatropica.2007.09.005
- Seidahmed OME, Hassan SA, Soghaier MA, et al. Spatial and temporal patterns of dengue transmission along a red sea coastline: a longitudinal entomological and serological survey in port Sudan city. PLoS Negl Trop Dis. 2012;6(9):e1821. doi:10.1371/journal.pntd.0001821
- Elduma AH, Osman WM. Dengue and hepatitis E virus infection in pregnant women in Eastern Sudan, A challenge for diagnosis in an endemic area. Pan Afr Med J. 2014;19:2–5. doi:10.11604/pamj.2014.19.391.5439
- Soghaier MA, Himatt S, Osman KE, et al. Cross-sectional community-based study of the socio-demographic factors associated with the prevalence of dengue in the eastern part of Sudan in 2011 infectious disease epidemiology. BMC Public Health. 2015;15(1):1–6. doi:10.1186/s12889-015-1913-0
- Teixeira MG. Few characteristics of dengue’s fever epidemiology in Brazil. Rev Inst Med Trop Sao Paulo. 2012;54(SUPPL.18):2–4. doi:10.1590/S0036-46652012000700002
- Mizuno Y, Kato Y, Kano S, Takasaki T. Imported malaria and dengue fever in returned travelers in Japan from 2005 to 2010. Travel Med Infect Dis. 2012;10(2):86–91. doi:10.1016/j.tmaid.2012.02.005
- Tuiskunen A, Monteil V, Plumet S, et al. Phenotypic and genotypic characterization of dengue virus isolates differentiates dengue fever and dengue hemorrhagic fever from dengue shock syndrome. Arch Virol. 2011;156(11):2023. doi:10.1007/s00705-011-1100-2
- Hamid Z, Hamid T, Alsedig K, et al. Molecular investigation of dengue virus serotype 2 circulation in Kassala state, Sudan. Jpn J Infect Dis. 2019;72(1):58–61. doi:10.7883/yoken.JJID.2018.267
- Abozeid S, Elsayed A, Schaffner F, Samy A. Re-emergence of Aedes aegypti in Egypt. Lancet Infect Dis. 2018;18:142–143. doi:10.1016/S1473-3099(18)30018-5
- Heikal OM, El-Bahnasawy MM, Morsy ATA, Khalil HHM. Aedes aegypti re-emerging in Egypt: a review and what should be done? J Egypt Soc Parasitol. 2011;41(3):801–814.
