Abstract
Background
As high touch wearable devices, the potential for microbial contamination of smart watches is high. In this study, microbial contamination of smart watches of healthcare workers (HCWs) was assessed and compared to the individual’s mobile phone and hands.
Methods
This study was part of a larger point prevalence survey of microbial contamination of mobile phones of HCWs at the emergency unit of a tertiary care facility. Swabs from smart watches, mobile phones and hands were obtained from four HCWs with dual ownership of these digital devices. Bacterial culture was carried out for all samples and those from smart watches and mobile phones were further assessed using shotgun metagenomic sequencing.
Results
Majority of the participants were females (n/N = 3/4; 75%). Although they all use their digital devices at work and believe that these devices could harbour microbes, cleaning in the preceding 24 hours was reported by one individual. Predominant organisms identified on bacterial culture were multidrug resistant Staphylococcus hominis and Staphylococcus epidermidis. At least one organism identified from the hands was also detected on all mobile phones and two smart watches. Shotgun metagenomics analysis demonstrated greater microbial number and diversity on mobile phones compared to smart watches. All devices had high signatures of Pseudomonas aeruginosa and associated bacteriophages and antibiotic resistance genes. Almost half of the antibiotic resistance genes (n/N = 35/75;46.6%) were present on all devices and majority were related to efflux pumps. Of the 201 virulence factor genes (VFG) identified, majority (n/N = 148/201;73%) were associated with P. aeruginosa with 96% (n/N = 142/148) present on smart watches and mobile phones.
Conclusion
This first report on microbial contamination of smart watches using metagenomics next generation sequencing showed similar pattern of contamination with microbes, VFG and antibiotic resistance genes across digital devices. Further studies on microbial contamination of wearable digital devices are urgently needed.
Introduction
Healthcare associated infections (HAI) associated with multidrug resistant pathogens remain a major health concern globally.Citation1,Citation2 Healthcare workers (HCWs) may inadvertently act as vectors in the transmission of these dangerous pathogens within the hospital setting and into the wider community. Studies using direct culture methods have demonstrated a high occurrence of microbial contamination of mobile phones of HCWs.Citation3–7 More recently studies using microbial sequencing techniques including 16s rRNA and Shotgun metagenomics have demonstrated a larger repertoire of microbes as well detection of virulence and antimicrobial resistance genes on mobile phones of HCWs.Citation8–11 This is within the context of high utilization and limited cleaning of these devices by HCWs in the workplace.Citation8,Citation12 These findings support the hypothesis that these high touch surfaces represent “trojan horses” contributing to the dissemination of pathogens.Citation4
In today’s technologically driven society, the use of smart watches is fast becoming as ubiquitous as mobile phones. The health and fitness applications available on these smart watches have increased their popularity in recent years as individuals strive towards attaining a healthy lifestyle. Similar to mobile phones, these smart watches are application (Apps) rich devices used for communication and also represent high touch surfaces. Furthermore, as they are wearable devices, they are in close contact with the skin thus interacting with the skin microbiome and are always carried from place to place as the wearer moves around. A bacterial culture-based study carried out in the United Kingdom before the advent of smart watches had shown that wearing a wristwatch is associated with increased bacterial contamination of the wrist and manipulation of the watch is associated with increased hand contamination.Citation13 More recently, bacterial culture of swabs of digital wristwatches of HCWs in a healthcare facility in India revealed the occurrence of bacterial contamination including detection of drug resistant isolates.Citation14 Hence the potential for microbial contamination of these devices and their role in the dissemination of microbes could be even more significant in comparison to mobile phones. Furthermore, the link between the microbial contamination of the smart watches and mobile phones is yet to be investigated. To address this gap in the literature, we present the first report on microbial contamination of smart watches of HCWs using shotgun metagenomic sequencing approach as well as the comparison of patterns of contamination detected on their mobile phones.
Materials and Methods
Study Site
This study was carried out at the Emergency care unit of Rashid Hospital, Dubai, United Arab Emirates, as part a larger study investigating microbial contamination of mobile phones among HCWs.Citation8 Whilst we had previously published findings on the microbial contamination of mobile phones of 35 HCWs at this facility,Citation8 the data obtained from HCWs with dual ownership of smart watches and mobile phones was not included in that report as additional sampling was carried out for these individuals. The Ethical approval was obtained from Dubai Scientific Research Ethics Committee, Dubai Health Authority (REF#: DSREC-02/2021_02) and MBRU Institutional Review Board (REF#: MBRU-IRB-2020-040) and consent was obtained from participants as per reported protocol.Citation8 The study was carried out in compliance with the Declaration of Helsinki.
Sample Collection
As previously described,Citation8 the study was designed as a point prevalence survey and therefore a single visit to the Emergency unit Rashid Hospital during the busy morning shift was carried out in April 2021 for the sampling. All the HCWs on duty at the time of the visit were invited to participate. No prior notice about the visit was provided to HCWs.Citation8 Four HCWs with dual ownership of mobile phones and smartwatches were identified during this sampling. For these individuals, in addition to swabs from their mobile phones, we obtained swab samples from their smart watches and both hands. Gloves were worn during the handling and swabbing of the digital devices and hands of these individuals. To prevent cross-contamination, gloves were changed between all swabbings. Sample collection was carried out using swabs pre-moistened with sterile normal saline and adequate contact of the applicator tip with the surfaces of the devices and hands during sampling. Two swabs were obtained for each device. The first swab sample for downstream next generation sequencing was placed in Zymos DNA/RNA Shield Collection Tubes (Zymos Research, Irvine CA, USA) and the second sample which was for bacterial culture was placed in Culture Swab EZ IITM” (Becton Dickinson) tubes. All smart watches and mobile phones sampled were flat screened without keypads. One swab sample for bacterial culture was obtained from the hands of the HCWs. Questionnaires were administered to obtain demographic profile as well as data on usage and cleaning of mobile phones during the work shift.
Bacterial Culture
This was carried out as previously described.Citation8 Briefly, swabs were incubated in sterile brain heart infusion (BHI) broth for 24–48 hours, followed by culture on blood agar, mannitol salt agar and McConkey agar plates. Bacterial identification and antibiotic susceptibility testing were carried out using the Vitek automated method (bioMerieux Marcy L’Etoile France). This study was carried out during the COVID-19 pandemic when heightened awareness of hand hygiene was in place, and as previously describedCitation8 the culture approach used ensured detection of low biomass of microbial contaminants, which might occur due to the prevailing circumstances.
Shotgun Metagenomics Sequencing
This was carried out at CosmosID Laboratories (Rockville, MD). Briefly, DNA extraction was carried out using the QIAmp® Powersoil Pro kit (Qiagen, Hilden, Germany) and isolated DNA was quantified by Qubit (ThermoFisher, Waltham MA, USA). Library construct, preparation and indexing were carried out as previously reported.Citation8 Sequencing was carried out on the Illumina HiSeq 4000 platform 2×150bp (Illumina). The CosmosID bioinformatics platform was used for the multi-kingdom microbiome analysis, profiling of antibiotic resistance and virulence genes and quantification of organisms’ relative abundance.Citation8,Citation9 Microbial “Richness” corresponds to the cumulative amount of all distinct microbes detected across all smart watches or mobile phones whereas the number of occurrences across these digital devices for each of these distinct microbes are represented by Hits. Metagenomics Sequence data are available on NCBI (https://www.ncbi.nlm.nih.gov/genbank/ BioProject ID: PRJNA750471).
Results
At the sampling time, all four HCWs owning both smart watches and mobile phones participated in the study. The majority were in the 26–55 years age group with a preponderance of females (). All four of them use their personal digital devices at work. Although all four HCWs indicated knowing that their digital devices could harbour microbes, cleaning of devices in the preceding 24 hours had been done by only one individual. Demographic data as well as reported usage and cleaning practices of the digital devices are found in .
Table 1 Demographic Profile and the Mobile Phone Utilization and Cleaning Practices of Participants
The microbes detected on swabs following bacterial culture are shown in with a predominance of multidrug resistant Staphylococcus hominis and Staphylococcus epidermidis. For all four HCWs, at least one organism identified on the hand swab was also detected on the mobile phone and this was true for the smart watches of two participants. For two participants, Micrococcus luteus and Staphylococcus epidermidis which were detected on their smart watches were not present either on their hands or mobile phones.
Table 2 Distribution of Bacteria Identified by Culture-Based Methods on Swabs Obtained from the Devices and Hands of Healthcare Workers
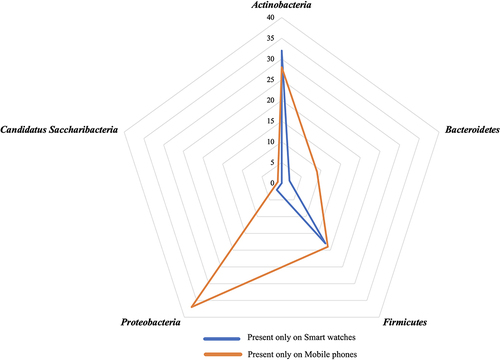
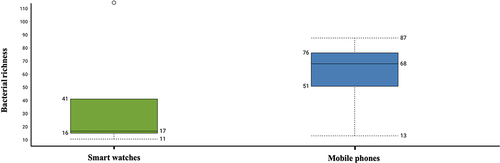
Shotgun metagenomics analysis showed that the average sequencing reads were 32 million and 45.3 million from mobile phones and smart watches respectively. Across all four mobile phones and four smartwatches, the richness of 223 different bacterial strains were found () and corresponded to a total occurrence across all digital devices of 394 hits (235 [60%] hits on mobile phones and 159 [40%] hits on smart watches) (). Fifty-four different bacterial strains were exclusively present on smart watches but absent on mobile phones (). Ninety-four bacterial strains were exclusively present on mobile phones but absent on smart watches and these were mainly associated with proteobacteria ().
Table 3 Abundance of Microbial Diversity and Number of Genes Related to Antibiotic Resistance and Virulent Factors for Each Participant
Figure 1 Boxplot showing the distribution of bacterial strains identified on smartwatches and mobile phones.
Predominant organisms (> 3/4 of phones; hits > 75%) found on mobile phones were generally greater in number and diversity when compared to organisms found on smart watches with the exception of Ralstonia pickettii and Propionibacterium spp (found in only 50% and 25% of mobile phones respectively) (). All organisms with high occurrence hits (> 75%) in mobile phones () were not entirely correlated with high occurrence hits in smart watches. Nine of the highest hit organisms present on mobile phones were found on only 25% of smart watches (). However, Aquabacterium parvum, Pseudomonas aeruginosa and two other pseudomonas species were present on all 4 mobile phones and 4 smart watches. S. aureus was found in three mobile phones and only one smart watch (the mobile phone of this smart watch owner was also contaminated with S. aureus). Of note, S. hominis, S. epidermidis, S. capitis were all detected on both the smart watch and mobile phone of a single HCW. S. cohnii was detected once from the swab of a single smart watch.
Figure 3 Comparison of bacteria present on smart watches and mobile phones.
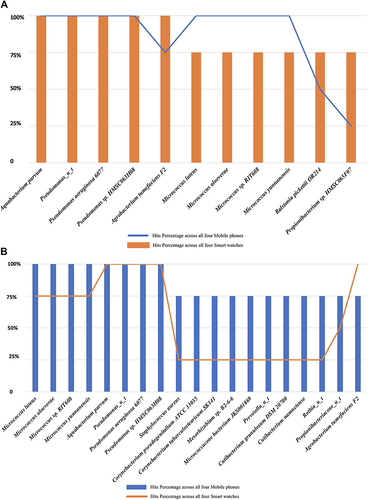
Additionally, HACEK bacteria including Haemophilus parahaemolyticus and H. parainfluenzae were found predominantly on mobile phones. Haemophilus parahaemolyticus was detected in only one smart watch while Cardiobacterium hominis was present on only one mobile phone. Acinetobacter baumannii was detected in both devices of the same HCW owner. With regards to fungi, Basidiomycota specifically Malassezia globosa and M. restricta were omnipresent in all smart watches and mobile phones.
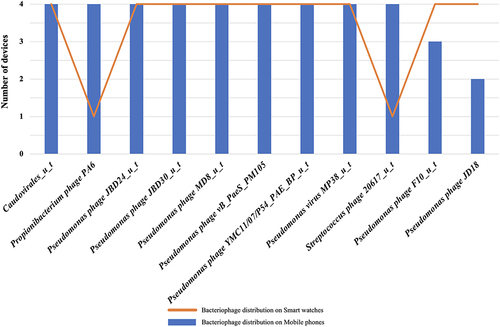
A total of 121 different strains of bacteriophages were found across all smart watches and mobile phones. shows the distribution of bacteriophages with the most predominant hits. A total of 201 distinct virulent factor genes were detected across all smart watches and mobile phones. Cumulative hits of virulent factors on smart watches were greater (but not statistically significant) than on mobile phones with 618 and 601 virulence factor genes (VFGs) respectively (). Thirty-two virulent factor genes were exclusively found on smart watches and not on phones while only 12 virulent factor genes were present exclusively on mobile phones (Supplementary Figure 1). Majority of the distinct VFGs (n/N = 148/201; 73%) were virulent genes associated with P. aeruginosa and most of these (n/N = 142/148) were present on all smart watches and all mobile phones. Of the remaining six, four virulent genes (orfA, orfC, tnpA, tnpR) were present only on smart watches while intl1 and GI3342496 were found only on mobile phones. Virulent genes present on all phone and watch devices included proteases inducing host immune-deregulations (aprA gene) as well as exoenzymes secreted by the type III secretion pathway (with the ubiquitous pathogenic exoT and exoY genes). Additionally, genes from Pseudomonas aeruginosa were associated with biofilm formation with alginate biosynthesis genes present in all mobile phones (4/4) and smartwatches (4/4). Four alginate switching genes were found and included algT, mucA, mucB, mucC. Moreover, five alginate response regulator genes were also present (algB, algR, algQ, algP, algZ) as well as 13 structural/biosynthesis alginate genes (algD, algX, algK, algE, algG, algL, algI, algJ, algF, algA, alg8, alg44 and algC).
Of the 75 antibiotic resistance genes detected, there was predominance of those encoding for multidrug resistance efflux pumps (n/N = 37/75; 49.3%). Antibiotic resistance genes for aminoglycosides (n/N = 13/75; 17.3%), beta lactams (n/N = 7/75; 9.3%) and macrolides (n/N = 8/75; 10.7%) were also present along with resistance genes for bleomycin, polymyxin, phenicols and tetracyclines. Of all the antibiotic resistant genes detected, 46.6% (n/N = 35/75) were present on all the smart watches and mobile phones. Supplementary Figure 2 shows the heatmap map distribution of antibiotic resistant genes in smart watches versus mobile phones. Among these 46.6% (n/N = 35/75) antibiotic resistant genes found on all digital devices, 88% (n/N = 31/35) were exclusively associated with multidrug efflux pumps. shows the list of multidrug resistance efflux pumps genes omnipresent on all smart watches and mobile phones as well as the nomenclature of their corresponding efflux pump phenotypes. The four other resistant genes not associated with efflux pumps and omnipresent on all eight devices were the Phenicol catB7 gene, the Aminoglycoside aph3’ IIb gene and two beta lactam resistant genes (beta lactamase blaOXA-50 and the Pseudomonas-derived cephalosporinase blaPDC2).
Table 4 Multidrug Resistance Efflux Pumps Genes Omnipresent on All Smart Watches and Mobile Phones with Reported Corresponding Efflux Pump Phenotypes
Additionally, 6 other efflux pumps related genes, not omnipresent to all 8 devices, were also found (of note, one smartwatch alone was able to harbour all of these 6 genes). These genes included msrE gene (belonging to the ABC-F ATP binding cassette efflux pump family), the Repressor-of-transcription mecI, the MDR-Efflux-pump qacA gene (belonging to the multidrug facilitator superfamily: MFS) found on both devices of the same owner, the Signal-transducing-protein mecR1cmx gene (Plasmid-or-transposon-encoded-chloramphenicol-exporter) found on one mobile phone and one smart watch of different individuals, the Repressor-of-efflux-complex phoP present on two smart watches and finally the gene Plasmid-or-transposon-encoded-chloramphenicol-exporter cmx which was detected on one smart watch. Metagenomic results also revealed the presence of additional genes involved in the regulation of efflux pumps on mobile phones and smart watches of participants ().
Table 5 Regulatory Genes for Efflux Pumps Present on Smart Watches and Mobile Phones of Healthcare Workers
Discussion
In this report, we present the first description of the microbial contamination of the hands, smart watches and mobile phones of HCWs with dual ownership of these digital devices. Findings from previous studies have shown that the culture-based approach does not demonstrate the extensive repertoire of microbial contamination on digital devices that are detectable with use of high-throughput next generation sequencing-based approaches.Citation8–10 Therefore, in this study, shotgun metagenomics sequencing of the swabs obtained from the smart watches and mobile phones of the HCWs was carried out to assess the spectrum of microbial contaminants as well as to detect the presence of virulence and antimicrobial resistance genes on these digital devices. In keeping with previous reports, the microbial identification and profiling from the metagenomic sequencing analysis were significantly more extensive compared to findings from the culture-based approach. Although the culture-based approach has the advantage of demonstrating microbial viability, the ability to identify only few culturable microbes remains a limitation.Citation13,Citation15,Citation16
In this study, findings from the culture-based approach showed similar bacteria being retrieved from the hands, smart watches and mobile phones of the same individual, an observation seen in 75% of the participants with the preponderant presence of Staphylococcus hominis and Staphylococcus epidermidis. This finding is in keeping with a recent report of staphylococci spp. being detected from the palmar surfaces and corresponding mobile phones of individuals in an urban community in Mexico.Citation17 Other prevalent bacteria identified from culture include Aerococcus viridans, Micrococcus luteus, Staphylococcus cohnii and Enterococcus faecalis. In addition, antimicrobial susceptibility testing revealed that the S. hominis, S. epidermidis and E. faecalis isolates exhibited a multidrug resistant phenotype. This is of concern as Staphylococcus spp and Enterococcus spp are often present in hospital settings, with the possibility of co-colonization of patients and they contribute to occurrence of nosocomial infections.Citation18,Citation19 Indeed, in a previous report on the acquisition of nosocomial pathogens after contact with environmental surfaces near patients, S. aureus and vancomycin resistant Enterococcus were the most predominant contaminants identified.Citation20 Therefore, our identification of these pathogens on the hands, smart watches and mobile phones of HCWs supports the call for the inclusion of sanitization of these digital devices in infection control practices alongside current hand hygiene practices in the healthcare setting.
The shotgun metagenomic sequencing of the smart watches and mobile phones identified all the bacteria found using the culture-based approach. However, an array of additional bacteria was identified including a high signature of Pseudomonas aeruginosa which is in keeping with previously reported work on mobile phones of HCWs from the same facility.Citation8 The intrinsic nature of this microbe to acquire resistance is demonstrated by the co-detection of a diversity of related antimicrobial resistance genes. This included resistance genes to different classes of antibiotics and the resistome showed a preponderance of genes related to efflux pumps. The ubiquitous presence of P. aeruginosa alongside related bacteriophages and antibiotic resistance genes on the smart watches and mobile phones of HCWs is worrisome. This is also compounded by the detection of various HACEK microorganisms including Acinetobacter baumannii which was found on both devices of a single HCW. The presence of these pathogenic microorganisms on smart watches enhances the potential risk of dissemination because, these wearable devices move everywhere with the wearer and thus are potential vectors for the dissemination of microorganisms and their related genetic elements particularly in the healthcare setting.
The existence of a diversity of microbes harbouring an arsenal of virulence and antimicrobial resistance genes on digital devices and specifically on wearable devices as we have demonstrated is of global public health importance. Whilst awareness of the potential role of mobile phones as “Trojan horse” for microbes is growing, this is the first report highlighting a potential similar role for wearable devices.Citation4 This work was carried out during the COVID-19 pandemic when heightened awareness of hand hygiene and environmental cleaning protocols was in place. The detection of microbial contamination on these devices despite these prevailing circumstances is a concern. Therefore, sanitisation of smart watches and mobile phones should be considered as a crucial element in infection control as contamination of these high touch devices potentially negate the efficacy of current hand hygiene protocols. This is because these live saving hand hygiene practices or even use of gloves are defeated if these “clean” or “gloved” hands are then immediately used to handle contaminated smart watches and mobile phones. Beyond the healthcare setting, this threat extends into the communityCitation17 and should be considered a potential biosecurity concern as global travel with modern transport facilitates the ease of movement of these devices (along with their microbial contaminants) across international borders.
Conclusion
We present the first study demonstrating the link in the microbial contamination of hands, smart watches and mobile phones of HCWs with dual ownership of these digital devices. Whilst the small number of participants in this study is a limitation, the findings highlight the need for further studies on microbial contamination of wearable devices particularly in the healthcare setting. In particular, such large studies will enable clustering analysis to determine the relationships between HCWs and the microbial contaminants on their digital devices. In addition, studies to investigate the efficacy ultra-violet C sanitizers for digital devices and the implementation of guidelines for their utilization in the hospital and community settings are urgently needed.
Data Sharing Statement
The authors confirm that the data supporting the findings of this study are available within the article. Metagenomics sequence data are available on NCBI (https://www.ncbi.nlm.nih.gov/genbank/ BioProject ID: PRJNA750471).
Disclosure
The authors report no conflicts of interest in this work.
Additional information
Funding
References
- Murray CJ, Ikuta KS, Sharara F. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi:10.1016/S0140-6736(21)02724-0
- van Duin D, Paterson DL. Multidrug-resistant bacteria in the community: trends and lessons learned. Infect Dis Clin North Am. 2016;30(2):377–390. doi:10.1016/j.idc.2016.02.004
- Saka KH, Akanbi AA, Obasa TO, et al. Bacterial contamination of hospital surfaces according to material make, last time of contact and last time of cleaning/disinfection. J Bacteriol Parasitol. 2017;8:3. doi:10.4172/2155-9597.1000312
- Olsen M, Campos M, Lohning A, et al. Mobile phones represent a pathway for microbial transmission: a scoping review. Travel Med Infect Dis. 2020;35:101704. doi:10.1016/j.tmaid.2020.101704
- Pillet S, Berthelot P, Gagneux-Brunon A, et al. Contamination of healthcare workers’ mobile phones by epidemic viruses. Clin Microbiol Infect. 2016;22(5):456 e451–456.
- Qureshi NQ, Mufarrih SH, Irfan S, et al. Mobile phones in the orthopedic operating room: microbial colonization and antimicrobial resistance. World J Orthop. 2020;11(5):252–264. doi:10.5312/wjo.v11.i5.252
- Selim HS, Abaza AF. Microbial contamination of mobile phones in a health care setting in Alexandria, Egypt. GMS Hyg Infect Control. 2015;10:Doc03. doi:10.3205/dgkh000246
- Boucherabine S, Nassar R, Zaher S, et al. Metagenomic sequencing and reverse transcriptase PCR reveal that mobile phones and environmental surfaces are reservoirs of multidrug-resistant superbugs and SARS-CoV-2. Front Cell Infect Microbiol. 2022;12:806077. doi:10.3389/fcimb.2022.806077
- Olsen M, Nassar R, Senok A, et al. A pilot metagenomic study reveals that community derived mobile phones are reservoirs of viable pathogenic microbes. Sci Rep. 2021;11(1):14102. doi:10.1038/s41598-021-93622-w
- Simmonds R, Lee D, Hayhurst E. Mobile phones as fomites for potential pathogens in hospitals: microbiome analysis reveals hidden contaminants. J Hosp Infect. 2020;104(2):207–213. doi:10.1016/j.jhin.2019.09.010
- Olsen M, Nassar R, Senok A, et al. Mobile phones are hazardous microbial platforms warranting robust public health and biosecurity protocols. Sci Rep. 2022;12:10009. doi:10.1038/s41598-022-14118-9
- Tajouri L, Campos M, Olsen M, et al. The role of mobile phones as a possible pathway for pathogen movement, a cross-sectional microbial analysis. Travel Med Infect Dis. 2021;43:102095. doi:10.1016/j.tmaid.2021.102095
- Jeans AR, Moore J, Nicol C, Bates C, Read RC. Wristwatch use and hospital-acquired infection. J Hosp Infect. 2010;74(1):16–21. doi:10.1016/j.jhin.2009.06.032
- Bhalla M, Aggarwal A, Fatima KH. Carbapenem-resistant bacteria on hand-held and hands-free electronic devices of healthcare workers and non-healthcare workers in Delhi, India. Infect Prev Pract. 2021;3(3):100162. doi:10.1016/j.infpip.2021.100162
- Cavari Y, Kaplan O, Zander A, Hazan G, Shemer-Avni Y, Borer A. Healthcare workers mobile phone usage: a potential risk for viral contamination. Surveillance pilot study. Infect Dis. 2016;48(6):432–435. doi:10.3109/23744235.2015.1133926
- Jansen AS, Balbinot GC, Daur AV, et al. Detection of potentially pathogenic bacteria on cell phones of hospital and university-based populations in Curitiba, southern Brazil. A cross-sectional study. Sao Paulo Med J. 2019;137(4):343–348. doi:10.1590/1516-3180.2018.044305072019
- Campista-León S, López-Espinoza JU, Garcia-Guerrero JT, Alfonso-Corrado C, Clark-Tapia R, Peinado-Guevara LI. Determination of drug-resistant bacteria in palmar surface and touchscreen cell phones from bystanders in an urban community. Microbiol Res. 2022;256:126958. doi:10.1016/j.micres.2021.126958
- Czekaj T, Ciszewski M, Szewczyk EM. Staphylococcus haemolyticus – an emerging threat in the twilight of the antibiotics age. Microbiol-SGM. 2015;161(11):2061–2068. doi:10.1099/mic.0.000178
- Sharaf EJ, Senok AC, Udo EE, Botta GA. Trafficking of methicillin-resistant staphylococci and co-colonization with vancomycin-resistant enterococci. Med Princ Pract. 2011;20(3):253–258. doi:10.1159/000323598
- Bhalla A, Pultz NJ, Gries DM, et al. Acquisition of nosocomial pathogens on hands after contact with environmental surfaces near hospitalized patients. Infect Control Hosp Epidemiol. 2004;25(2):164–167. doi:10.1086/502369