Abstract
Objective
The most common extraintestinal pathogen and infection site is uropathogenic Escherichia coli (UPEC), which causes urinary tract infections (UTIs). UPEC is also a common pathogen in bloodstream infections; in severe cases, it can lead to death. Although host and bacterial virulence factors have been demonstrated to be associated with UTI pathogenesis, the role of the related contributing factors in UTI and urinary source bacteremia is not yet fully understood. This study aimed to compare and analyze the factors contributing to urinary bacteremia in patients with UTI.
Methods
A total of 171 E. coli strains collected from patients with UTI and urinary source bacteremia at Chiayi Christian Hospital were used. Phylogenetic groups and virulence factors were determined using PCR. Drug resistance patterns were determined using the disk diffusion assay.
Results
Previous studies have demonstrated that fimbriae and papGII may be associated with first-step infections and severe UTIs, respectively. As expected, highly virulent E. coli strains (belonging to the phylogenetic B2 and D groups) were dominant in the bacteremic UTI (90%) and UTI (86.27%) groups. However, our results showed that the UTI group had a significantly higher prevalence of sfa/focDE (belonging to the S and FIC fimbriae) than the bacteremic UTI group (29.4% vs 12.5%; p=0.008). In the bacteremic group, we found that sfa/focDE was only detected in highly virulent strains. The bacteremic UTI group had a significantly higher prevalence of papGII (belonging to P fimbriae) than the UTI group (55.8% vs 37.3%; p=0.026). In addition, the P fimbriae gene cluster, including papC, papEF, and papGII, was predominant in highly virulent strains. Notably, our results show that multidrug-resistant (MDR) strains were significantly less virulent than non MDR strains.
Conclusion
Taken together, our results provide insights into the contributing factors in patients with UTI and urinary bacteremia.
Introduction
Urinary tract infections (UTI) are among the most common extra-intestinal infections caused by several pathogens.Citation1 Epidemiological studies have reported that various gram-positive and gram-negative bacteria, including Escherichia coli, Klebsiella pneumonia, Enterococcus faecalis, Proteus mirabilis, Pseudomonas aeruginosa, and Staphylococcus saprophyticus can cause UTIs.Citation2–4 The leading UTI-causing pathogen, E. coli, can cause severe invasive diseases such as acute kidney injury, bacteremia, and sepsis.Citation2,Citation5–7 Approximately 30% of sepsis cases originating from urinary sites are caused by malignancies of the urinary tract.Citation6 Notably, the all-cause mortality rate of sepsis ranges from 28–56%.Citation6 Therefore, an investigation of the factors that contribute to the pathogenesis of sepsis is urgently needed.
Several factors, including virulence and antibiotic resistance genes in uropathogenic E. coli (UPEC), could affect the severity of UTIs.Citation8,Citation9 Virulence genes are key factors that assist UPEC in host invasion, colonization, and survival in the host.Citation9,Citation10 In addition, UPEC can be further divided into different phylogenetic groups that differ in their gene content, pathogenicity islands, virulence factors, and genomic islands, resulting in great diversity in the pathogenicity and antibiotic resistance rates in bacteria.Citation9,Citation11,Citation12 The adherence of pathogens to host cells is a key step in the pathogenesis.Citation13,Citation14 Several adhesins, including type I, type II, S, and FIC fimbriae, are involved in host cell invasion.Citation15 Type I fimbriae are regulated by fim gene clusters and are related to biofilm formation on abiotic surfaces and binding to kidney cells.Citation16 Type II fimbriae are encoded by pap genes required for P-fimbrial synthesis.Citation17,Citation18 Previous studies reported that the papGII gene plays a vital role in developing bacteremia in patients who have upper UTI.Citation17 The expression of type I and P fimbriae is controlled by phase variations in E. coli.Citation19,Citation20 Type I fimbriae are repressed during P fimbrial expression and vice versa.Citation8 Although multiple virulence factors have been reported in the pathogenesis of UTIs, the physiological role of virulence factors associated with invasiveness remain incompletely defined.Citation21
The aim of this study was two-fold. First, we estimated the prevalence of virulence factors in UTI and urinary-source bacteremia isolates associated with bacterial pathogenesis and patient progression. Second, we determined the prevalence of drug resistance patterns in the same sample. In conclusion, this study provides information concerning the factors contributing to urinary bacteremia in pathogens and patients with UTI.
Material and Methods
Study Setting
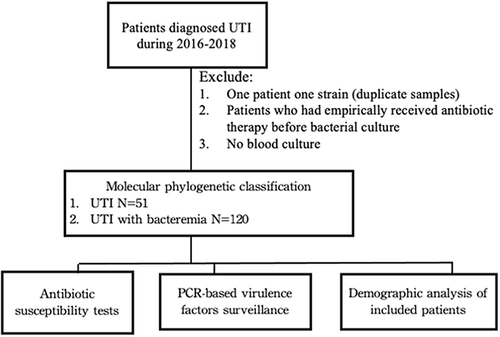
Patients hospitalized with a diagnosis of UTI or urinary bacteremia between 2016 and 2018 were enrolled in this study. Bacterial strains were isolated for phylogenetic classification and virulence factor determination. Patients who received antibiotic therapy before bacterial culture were excluded. Moreover, patients with UTI were excluded from the study because they had no blood cultures (). Demographic characteristics and clinical features of the patients were also studied.
Molecular Phylogenetic Classification
Bacterial genomic DNA was extracted according to the manufacturer’s instructions (QIAGEN). Escherichia coli phylogenetic groups were divided into four groups (A, B1, B2, and D) based on specific genetic markers (chuA, yjaA, and TSPE4.C2) using PCR, as described by Cermont et al.Citation22
Virulence Factors Determination
Extracted bacterial DNA was used to detect 26 virulence genes. Virulence genes, including adhesins (1. P fimbriae: papAH, papC, papEF, papGI, papGII; 2. M fimbriae: bmaE; 3. S and FIC fimbriae: sfa/focDE, focG, sfaS; 4. Type I fimbriae: fimH; 5. Dr binding adhesin: afa/draBC; 6.N-acyl D-glucosamine specific fimbriae: gafD; 6. Non-fimbriae adhesin:nfaE), capsules (kpsMTII, kpsMTIII), iron acquisition system (fyuA, iuTA), toxins (hylA, cnf1, cdtB-A, cdtB-S), and vasin and protein genes (cvaT, ibeA,traT, rfc) were detected using PCR using specific primer pairs as described in previous studies.Citation12,Citation23,Citation24 Positive and negative controls were used for each PCR assay.
Antibiotic Susceptibility Test
The antibiotic susceptibility was performed using disc diffusion assay against 13 antibiotics (beta-lactam: ampicillin/sulbactam, piperacillin/tazobactam; aminoglycosides: amikacin, gentamicin; 1st–2nd cephalosporins: cefazolin, cefuroxime; 3rd–4th generation cephalosporin: cefotaxime, cefepime; Carbapenem: ertapenem, meropenem; fluoroquinolone: levofloxacin; folate pathway/sulfamethoxazole: trimethoprim/sulfamethoxazole [TMP/SMX]), according to the recommendations of Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2020). Quality control strains, E. coli ATCC25922 and Pseudomonas aeruginosa ATCC2785, were used for the tests.
Statistical Analysis
All statistical analyses were performed using SPSS statistical software for Windows version 23 (IBM Corporation, Armonk, NY, USA). Pearson’s chi-square, Fisher’s exact, and Mann–Whitney U-tests were used to compare variables between the different groups. Virulence scores were calculated as the sum of the virulence factors in each isolate. Statistical significance was set at p < 0.05.
Results
Characteristics of Patients and Clinical Data
A total of 171 patients with UTI were enrolled in the study, of whom 120 (70.18%) were diagnosed with UTI with bacteremia and 51 (29.82%) without bacteremia. The most common underlying disease was hypertension (54.4%), followed by upper UTI (46.2%), and diabetes mellitus (45.6%) (). As shown in , no significant differences were observed in the clinical characteristics between the two groups, except for hypertension and septic shock, which showed that patients with bacteremia had a higher prevalence of hypertension (60% vs 41.2%; p=0.024) and septic shock (30.8% vs 9.8%; p=0.003) than patients without bacteremia. Notably, patients with UTI with bacteremia had a longer average length of hospital stay than patients with UTI without bacteremia (9.52 ± 4.14 vs 7.73 ± 3.46; p=0.007) ().
Table 1 Demographic Characteristics of the Study Patients and Isolated Strains
In this study, phylogenetic groups were grouped based on the presence or absence chuA, yjaA, and TSPE4-2. Of the 171 isolates, 118 (69%) belonged to group B2, followed by groups D (19.9%), A (5.8%), and B1 (5.3%). Phylogenetic groups B2 and D dominated in the two patient groups. The phylogenetic groups did not differ significantly between patients with UTI with and without bacteremia ().
Prevalence of Virulence Factors in UTI and Bacteremic UTI Groups
To determine the pathogenicity and association between virulence factors and phylogenetic groups (high vs low virulence) in patients with UTI and bacteremia, 26 virulence genes classified into 5 groups were examined and analyzed. The prevalence of virulence factors detected in isolated strains has been 93.6% for fimH, 85.4% for fyuA, 68.4% for iutA, 67.8% for traT and kpsMTII, 62.6% for papAH, 61.4% for papC,60.2% for papEF and 50.3% for papGIII (). Interestingly, the bacteremic UTI group had a significantly higher prevalence of papGII (P fimbriae) than the UTI group (55.8% vs 37.3%; p=0.026) (). However, the bacteremic UTI group had a significantly lower prevalence of sfa/focDE (S and FIC fimbriae) than the UTI group (12.5% vs 29.4%; p=0.008) (). The UTI group had a significantly higher prevalence of S and FIC fimbriae adhesins than the bacteremic UTI group (43.1% vs 24.2%; p=0.013) ().
Table 2 Prevalence of Virulent Factors Identified from Pathogenic Strains in UTI Infected Patients with or Without Bacteremia
Association among strain virulence, virulence factors, and demographic factors in patients with UTI with or without bacteremia
The prevalence of virulence factors in high (B2 and D strains) and low-virulence strains (A and B1 strains) isolated from patients with bacteremic UTI and UTI is presented in . As expected, highly virulent E. coli strains were dominant in patients with bacteremic UTI (90%) and UTI (86.27%) (). In contrast to low virulence group, high virulence group had significantly high prevalence of pap gene cluster including papC (bacteremic UTI: 70.4% vs 25.0%; p=0.03; UTI: 59.1% vs 0%; p=0.04), papEF (bacteremic UTI: 66.7% vs 33.3%; p=0.03; UTI: 59.1% vs 14.3%; p=0.042) and papGII (bacteremic UTI: 59.3% vs 25.0%; p=0.023) in both bacteremic UTI and UTI patient groups ().
Table 3 Prevalence of Virulent Factors Identified from High/Low Pathogenic Strains in UTI Infected Patients with or without Bacteremia
According to the virulence gene classification, the highly virulent bacteremic UTI group had a significantly higher prevalence of P fimbriae adhesin than the low-virulence bacteremic UTI group (78.7% vs 41.7%; p=0.01). In iron acquisition system related genes, high prevalence related genes were found in high virulent UTI group (bacteremic UTI: 92.6% vs 75.0%; p=0.08; UTI: 75.0 vs 28.6%; p=0.025) (). Notably, the highly virulent UTI group had a significantly higher prevalence of invasin and protectin genes than the low-virulence group (75% vs 28.6%; p=0.025), whereas no significant difference was observed in the bacteremic UTI group (74.1% vs 66.7%; p=0.731) ().
To determine the association between strain virulence and demographic factors, the Charlson Comorbidity Index (CCI), length of hospital stay, and related diseases were analyzed (). Patients with a high CCI were more likely to be in low virulent UTI strain group than in high virulent strain group (6.29 ± 2.69 vs 3.64 ± 2.76; p=0.022) (). Notably, the highly virulent UTI group had a significantly higher prevalence of upper UTI than the low-virulence UTI group (43.2% vs 0%, p=0.037) (). However, the upper UTI rate did not differ significantly between the high- and low-virulence bacteremic UTI patient groups (50.0% vs 50.0%, p=1.000) (). In addition, multidrug resistance (MDR) rates did not differ significantly between the bacteremic UTI and UTI patient groups (bacteremic UTI: 35.2% vs 33.3%; p=1.000; UTI: 50.0% vs 42.9%; p=1.000) ().
Virulence scores differed significantly between high- and low-virulence strains (). Highly virulent strains recovered from both blood and urine samples had significantly higher virulence scores, especially adhesin and iron acquisition system gene scores (). Notably, the highly virulent UTI group had significantly higher capsule gene scores than the low-virulence UTI strain group ().
Table 4 Comparison of Virulence Scores Calculated from High/Low Pathogenic Strains in UTI Infected Patients with or Without Bacteremia
Association among MDR, virulence scores, and demographic factors in patients with UTI with or without bacteremia
To analyze MDR and the association between virulence scores and demographic factors in patients, antibiotic susceptibility tests were performed. The results showed that all isolates were susceptible to ertapenem and meropenem. Notably, the isolated strains were more susceptible to amikacin (98.8%) and piperacillin/tazobactam (96.5%) but showed high rates of resistance to cefazolin (69.6%) and TMP/SMX (49.7%). Notably, no significant differences in drug resistance patterns were found between high and low-virulence strains.
Of the 171 isolates, 39.18% (67/171) were MDR strains. In contrast to the bacteremic UTI group, the UTI group had a higher prevalence of MDR (25/26, 49.02% vs 42/120, 35%) (). As shown in , the MDR rates in the different phylogenetic groups did not differ significantly between the bacteremic UTI and UTI patient groups (). To analyze the association between MDR and virulence scores, patients infected with non-MDR strains had significantly higher invasin and protectin gene score than those infected with MDR strains (0.97 ± 0.69 vs 0.72 ± 0.57; p=0.013) ().
Table 5 Characteristics of MDR and Non-MDR Strains Isolated from UTI Infected Patients with or Without Bacteremia
Notably, MDR strains isolated from patients with bacteremia had a significantly higher prevalence of acute kidney injury than non-MDR strains (AKD: 33.3% vs 6.4%, p<0.001) (). A similar trend was found in patients with upper UTI (61.9% vs 43.6%, p=0.056) (). Expectedly, patients with bacteremic UTI infected with MDR strains had significantly long average length of hospital stay days than those infected with non-MDR strains (bacteremic UTI: 10.69 ± 4.59 vs 8.88 ± 3.76; p=0.022; UTI: 8.76 ± 4.11 vs 6.73 ± 2.39; p=0.035) ().
Discussion
In this molecular epidemiological analysis of pathogenic E. coli clinical strains from patients with UTI and urinary source bacteremia, we found that virulence factor is associated with strain virulence and UTI disease progression. The crucially important first step during infection is adhesion of E. coli strain to urothelial cell.Citation25 Alternatively, pathogen’s ability to colonize and invade host cells strictly depend on adhesins such as fimbriae.Citation25,Citation26 Compared to bacteremic UTI group, E. coli isolated from UTI group with high prevalence of sfa/focDE gene (belonged to S and FIC fimbriae) was observed (29.4% vs 12.5%; p=0.008). Besides, we also found that UTI group had significantly higher prevalence of S and FIC fimbriae adhesins than bacteremic UTI group (43.1% vs 24.2%; p=0.013). In this study, our results suggested S and FIC fimbriae might play an important role in the first step of infection. Previous studies also demonstrated that S and FIC fimbriae are correlated with UPEC pathogenicity and mediate binding to kidney cell.Citation27–29
The role of adhesins in UTI to bacteremic UTI is not fully understood. In the bacteremic UTI group, we found that sfa/focDE gene was only detected from high virulent bacteremic group. Naziri et al showed that almost all (99%) of UPEC isolated had biofilm formation ability, whereas only presence of sfa/focDE gene was significantly association with moderate and strong biofilm formation.Citation30 The role of sfa/focDE gene in pathogenic strains associated with disease progression (UTI to bacteremic UTI) remains to be investigate experimentally. In addition, high virulent strain had a significantly higher prevalence of P fimbriae including papC, papEF and papGII (). Besides, our results showed that high virulent bacteremic UTI groups had significantly higher prevalence of papGII gene than low virulent group (). Recently, Biggel et al using genome-wide association approach to analyze 385 invasive UPEC isolates and 337 non-invasive UPEC isolates.Citation17 They reported papGII locus as the key features specifically associated with invasiveness and severe UTI.Citation17
Interestingly, we found no significant difference in virulence scores and prevalence of MDR between unclassified strains isolated from blood and urine (). Kim et al showed the similar trends by using whole genome sequencing technique to analyze 80 clinical strains.Citation31 However, virulence scores were significantly different between high virulent and low virulent groups. In general, high virulent strains had significantly high virulence scores than low virulent strains (). Besides, our results showed that both bacteremic and MDR strain groups has directly impacted extended hospital stays. The disease progression and severity associated with virulence factors needs to be confirmed experimentally.
Diabetes has been reported to be associated with UTI infection.Citation7,Citation32 In our study, high virulent strains were dominant at bacteremic UTI with diabetes (50.9%; 55/108). Over all, 96.5% (55/57) of bacteremic UTI patients with diabetes were infected with high virulent E. coli strains (B2:36, 63.2%; D:19, 33.3%). The similar trend was observed in patient with upper UTI. All upper UTI patients were infected with high virulent strains. In this study, high virulent strains have been found to be associated with pathogenesis of UTI in patients with diabetes or upper UTI.
In the past decade, an increase and spread of MDR UPEC strains has been observed and become a public health concern, particularly in women with recurrent UTIs.Citation21,Citation33 In the study, 39.18% MDR UPEC strains were isolated. In addition, no significant difference in the MDR UPEC isolation rates between high virulent and low virulent or bacteremic UTI and UTI groups (). Interestingly, MDR strains were significantly less virulent than non MDR strains (). However, large multiple-center cohort studies must be initiated to confirm the relationship between drug resistance and bacteria virulence. To adapt and survival in the host, the pathogen modulates its virulence potential and fitness.Citation34 Some studies reported that bacterial virulence and fitness are directly or indirectly affected by drug resistance.Citation35–37 Hwang et al reported that MDR strains were significantly less virulent than drug sensitive strains.Citation37 Pathogens occur in the presence of various selective pressures promote its evolution in a direction to increase the survival fitness.Citation38
This study has several limitations. First limitation of the present study was small sample size in certain subgroup, which reduced statistical power for detecting population differences. We hope to initiate a large-scale collection in multiple centers to further clarify association between virulence factors and host diseases. Second, it will be worth comparing the characteristics of isolated strains including prevalence of virulence factors and drug resistant pattern to those collect from the different region of hospital.
In summary, this study provided insight into the contributing factors in patients with UTI to urinary source bacteremia. Our results revealed that P fimbriae (including papC, papEF and papGII) and sfa/focDE might be associated with invasiveness and disease progression, respectively. These findings have implications for the pathogenesis and control measures in patients with UTI infection.
Ethical Statement
This study was approved by the Institutional Review Board of Ditmanson Medical Foundation Chia-Yi Christian Hospital (IRB NO: CYCH-106040). Informed consent was obtained from all study participants prior to study commencement. This study complied with the Declaration of Helsinki.
Disclosure
The authors declare they have no conflicts of interest.
Acknowledgments
This study was supported by grants from Ditmanson Medical Foundation Chia-Yi Christian Hospital Research Program (R106-028) and National Science Council (NSC 109-2314-B-415-002-MY3) (Taipei, Taiwan).
References
- Yi-Te C, Shigemura K, Nishimoto K. et al. Urinary tract infection pathogens and antimicrobial susceptibilities in Kobe, Japan and Taipei, Taiwan: an international analysis. J Int Med Res. 2020;48(2):300060519867826. doi:10.1177/0300060519867826
- Abraham SN, Miao Y. The nature of immune responses to urinary tract infections. Nat Rev Immunol. 2015;15(10):655–663. doi:10.1038/nri3887
- Salm J, Salm F, Arendarski P, Kramer TS. High frequency of enterococcus faecalis detected in urinary tract infections in male outpatients - a retrospective, multicenter analysis, Germany 2015 to 2020. BMC Infect Dis. 2023;23(1):812. doi:10.1186/s12879-023-08824-6
- Kao CY, Zhang YZ, Bregente CJB, et al. A 24-year longitudinal study of Klebsiella pneumoniae isolated from patients with bacteraemia and urinary tract infections reveals the association between capsular serotypes, antibiotic resistance, and virulence gene distribution. Epidemiol Infect. 2023:151:e155. 10.1017/S0950268823001486.
- Kao CY, Zhang YZ, Yang DC, et al. Characterization of host and Escherichia coli strains causing recurrent urinary tract infections based on molecular typing. BMC Microbiol. 2023;23(1):90. doi:10.1186/s12866-023-02820-1
- Barnett BJ, Stephens DS. Urinary tract infection: an overview. Am J Med Sci. 1997;314(4):245–249. doi:10.1097/00000441-199710000-00007
- Bonten M, Johnson JR, van den Biggelaar AHJ, et al. Epidemiology of Escherichia coli bacteremia: a systematic literature review. Clin Infect Dis. 2021;72(7):1211–1219. doi:10.1093/cid/ciaa210
- Naziri Z, Derakhshandeh A, Soltani Borchaloee A, Poormaleknia M, Azimzadeh N. Treatment failure in urinary tract infections: a warning witness for virulent multi-drug resistant ESBL- producing Escherichia coli. Infect Drug Resist. 2020;13:1839–1850. doi:10.2147/IDR.S256131
- Bunduki GK, Heinz E, Phiri VS, Noah P, Feasey N, Musaya J. Virulence factors and antimicrobial resistance of uropathogenic Escherichia coli (UPEC) isolated from urinary tract infections: a systematic review and meta-analysis. BMC Infect Dis. 2021;21(1):753. doi:10.1186/s12879-021-06435-7
- Hagan EC, Lloyd AL, Rasko DA, Faerber GJ, Mobley HL. Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS Pathog. 2010;6(11):e1001187. doi:10.1371/journal.ppat.1001187
- Rezatofighi SE, Mirzarazi M, Salehi M. Virulence genes and phylogenetic groups of uropathogenic Escherichia coli isolates from patients with urinary tract infection and uninfected control subjects: a case-control study. BMC Infect Dis. 2021;21(1):361. doi:10.1186/s12879-021-06036-4
- Yousefipour M, Rezatofighi SE, Ardakani MR. Detection and characterization of hybrid uropathogenic Escherichia coli strains among E. coli isolates causing community-acquired urinary tract infection. J Med Microbiol. 2023;72(2). doi:10.1099/jmm.0.001660
- Schuroff PA, Abe CM, Silva JW, et al. Role of aggregate-forming pilus (AFP) in adherence and colonization of both intestinal and urinary tracts. Virulence. 2022;13(1):1423–1433. doi:10.1080/21505594.2022.2112818
- Beerepoot M, Geerlings S. Non-antibiotic prophylaxis for urinary tract infections. Pathogens. 2016;5(2). doi:10.3390/pathogens5020036
- Nielubowicz GR, Mobley HL. Host-pathogen interactions in urinary tract infection. Nat Rev Urol. 2010;7(8):430–441. doi:10.1038/nrurol.2010.101
- Zuberi A, Ahmad N, Khan AU. CRISPRi induced suppression of fimbriae gene (fimH) of a uropathogenic Escherichia coli: An approach to inhibit microbial biofilms. Front Immunol. 2017;8:1552. doi:10.3389/fimmu.2017.01552
- Biggel M, Xavier BB, Johnson JR, et al. Horizontally acquired papGII-containing pathogenicity islands underlie the emergence of invasive uropathogenic Escherichia coli lineages. Nat Commun. 2020;11(1):5968. doi:10.1038/s41467-020-19714-9
- Cuenod A, Agnetti J, Seth-Smith HMB, et al. Bacterial genome-wide association study substantiates papGII of Escherichia coli as a major risk factor for urosepsis. Genome Med. 2023;15(1):89. doi:10.1186/s13073-023-01243-x
- Struve C, Krogfelt KA. In vivo detection of Escherichia coli type 1 fimbrial expression and phase variation during experimental urinary tract infection. Microbiology. 1999;145(Pt 10):2683–2690. doi:10.1099/00221287-145-10-2683
- Zamora M, Ziegler CA, Freddolino PL, Wolfe AJ. A thermosensitive, phase-variable epigenetic switch: pap revisited. Microbiol Mol Biol Rev. 2020;84(3):1.
- Mancuso G, Midiri A, Gerace E, Marra M, Zummo S, Biondo C. Urinary tract infections: the current scenario and future prospects. Pathogens. 2023;12(4). doi:10.3390/pathogens12040623
- Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66(10):4555–4558. doi:10.1128/AEM.66.10.4555-4558.2000
- Lin WH, Wang MC, Liu PY, et al. Escherichia coli urinary tract infections: host age-related differences in bacterial virulence factors and antimicrobial susceptibility. J Microbiol Immunol Infect. 2022;55(2):249–256. doi:10.1016/j.jmii.2021.04.001
- Rodriguez-Bano J, Mingorance J, Fernandez-Romero N, et al. Virulence profiles of bacteremic extended-spectrum beta-lactamase-producing Escherichia coli: association with epidemiological and clinical features. PLoS One. 2012;7(9):e44238. doi:10.1371/journal.pone.0044238
- Krachler AM, Orth K. Targeting the bacteria-host interface: strategies in anti-adhesion therapy. Virulence. 2013;4(4):284–294. doi:10.4161/viru.24606
- Pak J, Pu Y, Zhang ZT, Hasty DL, Wu XR. Tamm-Horsfall protein binds to type 1 fimbriated Escherichia coli and prevents E. coli from binding to uroplakin Ia and Ib receptors. J Biol Chem. 2001;276(13):9924–9930. doi:10.1074/jbc.M008610200
- Ejrnaes K. Bacterial characteristics of importance for recurrent urinary tract infections caused by Escherichia coli. Dan Med Bull Apr. 2011;58(4):1.
- Antao EM, Wieler LH, Ewers C. Adhesive threads of extraintestinal pathogenic Escherichia coli. Gut Pathog. 2009;1(1):22. doi:10.1186/1757-4749-1-22
- Khan AS, Kniep B, Oelschlaeger TA, Van Die I, Korhonen T, Hacker J. Receptor structure for F1C fimbriae of uropathogenic Escherichia coli. Infect Immun. 2000;68(6):3541–3547. doi:10.1128/IAI.68.6.3541-3547.2000
- Naziri Z, Kilegolan JA, Moezzi MS, Derakhshandeh A. Biofilm formation by uropathogenic Escherichia coli: a complicating factor for treatment and recurrence of urinary tract infections. J Hosp Infect. 2021;117:9–16. doi:10.1016/j.jhin.2021.08.017
- Kim B, Kim JH, Lee Y. Virulence factors associated with Escherichia coli bacteremia and urinary tract infection. Ann Lab Med. 2022;42(2):203–212. doi:10.3343/alm.2022.42.2.203
- Paudel S, John PP, Poorbaghi SL, Randis TM, Kulkarni R. Systematic review of literature examining bacterial urinary tract infections in diabetes. J Diabetes Res. 2022;2022:3588297. doi:10.1155/2022/3588297
- Walker MM, Roberts JA, Rogers BA, Harris PNA, Sime FB. Current and emerging treatment options for multidrug resistant Escherichia coli urosepsis: a review. Antibiotics (Basel). 2022;11(12):1821. doi:10.3390/antibiotics11121821
- Beceiro A, Tomas M, Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev Apr. 2013;26(2):185–230. doi:10.1128/CMR.00059-12
- Russo TA, MacDonald U, Beanan JM, et al. Penicillin-binding protein 7/8 contributes to the survival of Acinetobacter baumannii in vitro and in vivo. J Infect Dis. 2009;199(4):513–521. doi:10.1086/596317
- Cayo R, Rodriguez MC, Espinal P, et al. Analysis of genes encoding penicillin-binding proteins in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55(12):5907–5913. doi:10.1128/AAC.00459-11
- Hwang W, Yoon SS. Virulence characteristics and an action mode of antibiotic resistance in multidrug-resistant pseudomonas aeruginosa. Sci Rep. 2019;9(1):487. doi:10.1038/s41598-018-37422-9
- Diard M, Hardt WD. Evolution of bacterial virulence. FEMS Microbiol Rev. 2017;41(5):679–697. doi:10.1093/femsre/fux023