Abstract
Staphyloxanthin is a virulence factor which protects Staphylococcus aureus in stress conditions. We isolated two pigment variants of S. aureus and one strain of Pseudomonas aeruginosa from a single wound infection. S. aureus variants displayed white and yellow colony phenotypes. The sequence of the operons for staphyloxanthin synthesis indicated that coding and promoter regions were identical between the two pigment variants. Quorum sensing controls pigment synthesis in some bacteria. It is also shown that P. aeruginosa quorum-sensing molecules affect S. aureus transcription. We explored whether the co-infecting P. aeruginosa can affect pigment production in the white S. aureus variant. In co-culture experiments between the white variants and a selected number of Gram-positive and Gram-negative bacteria, only P. aeruginosa induced pigment production in the white variant. Gene expression analysis of the white variant did not indicate upregulation of the crtM and other genes known to be involved in pigment production (sigB, sarA, farnesyl pyrophosphate synthase gene [FPP-synthase], hfq). In contrast, transcription of the catalase gene was significantly upregulated after co-culture. P. aeruginosa-induced pigment synthesis and catalase upregulation correlated with increased resistance to polymyxin B, hydrogen peroxide, and the intracellular environment of macrophages. Our data indicate the presence of silent but functional staphyloxanthin synthesis machinery in a white phenotypic variant of S. aureus which is activated by a co-infecting P. aeruginosa via inter-species communication. Another S. aureus virulence factor, catalase is also induced by this co-infecting bacterium. The resulting phenotypic changes are directly correlated with resistance of the white variant to stressful conditions.
Introduction
More than 90% of all Staphylococcus aureus isolates from human infections are golden pigmented.Citation1 In 1884, RosenbachCitation2 classified different isolates of Staphylococci based on pigment production as S. aureus (pigmented) and S. albus (non-pigmented). This simple classification method was later replaced by more advanced classification using other properties of the bacterium.Citation3–Citation5 Although most isolates of S. aureus produce golden colonies, it was recognized that some Staphylococcus isolates, which displayed all characteristics of S. aureus, could produce white colonies, which was partly dependent on growth media.Citation6 Conclusive proof that these white colonies belonged to S. aureus lineage may have required modern molecular tools not available at the time. Pigment synthesis in S. aureus is carried out by the enzymes of the crtOPQMN operon, but it is not known whether the presence of an intact operon in the genome would inevitably lead to development of the stable golden colony phenotype. In a recent study, it was shown that a S. aureus isolate with a white-colony phenotype actually lacked the crtOPQMN operon.Citation7 However, these authors proposed to reclassify the isolate and the lineage it belonged to as S. argenteus.
Golden pigment in S. aureus is the manifestation of triterpenoid staphyloxanthin biosynthesis, which is transported to and embedded in the cellular membrane.Citation8,Citation9 Staphyloxanthin is synthesized through a five-step enzymatic process by the five enzymes of the polycistronic crtOPQMN operon which is controlled by a single promoter.Citation10 In this process, farnesyl diphosphate acts as the substrate for the initial step and staphyloxanthin is the product of the final step.Citation11
Staphyloxanthin has been considered one of the several virulence factors that S. aureus has at its disposal to withstand and survive a stressful environment. Clauditz et al have used targeted mutagenesis of the genes from the crtOPQMN operon to demonstrate the importance of staphyloxanthin for resistance of S. aureus to oxidative stress.Citation12 It was shown that wild type pigment-producing strains withstood exposure to higher concentrations of H2O2 than the isogenic, pigment knockout mutants.Citation12 Similarly, Liu et al reported that the golden pigment impairs neutrophil killing, promotes virulence due to its antioxidative action, and significantly contributes to the development of subcutaneous abscess in an animal model.Citation13
In addition to its antioxidative activity, staphyloxanthin also affects the chemical composition and functional activity of the S. aureus cellular membrane by reducing membrane fluidity and thereby stabilizing the membrane structure.Citation14 The change in membrane fluidity translates into higher resistance to host antimicrobial peptides used by the host to eradicate bacterial agents.Citation15 The enzymes of the staphyloxanthin biosynthetic pathway are considered prime targets for drug development aimed at eradication of S. aureus infections. The first enzyme in this pathway, CrtM, has a similar catalytic site structure to that of human squalene synthase which is involved in cholesterol biosynthesis. Liu et al showed that inhibitors of human squalene synthase could prevent pigment production in S. aureus, and affect its survival.Citation16 Two molecules, N-3-(3-phenoxyphenyl) propylphosphonoacetamide and phosphonosulfonate have been identified as CrtM inhibitors.Citation17 However, specificity and in vivo applicability of these molecules need further investigation.
Wound infection is a significant problem in clinical settings, particularly in combat-related wounds where improvised explosive devices not only damage tissue but also deposit dirt and metal fragments in wounds. S. aureus is commonly isolated from wound infections and has become an alarming problem due to the emergence of several methicillin-resistant strains (eg, methicillin resistant staphylococcus aureus [MRSA]).Citation18 Wound infections are polymicrobial and S. aureus is commonly isolated from these infections along with a consortium of other bacteria.Citation19–Citation22 At present, it is almost impossible to define the role of each participating bacterium of these communities in the outcome of infection.Citation23 On the other hand, to better understand the mechanisms of persistence and in vivo survival of S. aureus during infections, it is necessary to understand the interaction between S. aureus and its co-infecting bacteria. One of the co-infecting bacterial species which is routinely isolated from clinical samples along with S. aureus is Pseudomonas aeruginosa. Samples from wound infections and lungs of patients with cystic fibrosis often contains both P. aeruginosa and S. aureus, underscoring the clinical importance of co-infection by these two bacteria.Citation24,Citation25 It has been shown that interaction of P. aeruginosa and Gram-positive bacteria including S. aureus can lead to increased virulence in P. aeruginosa.Citation26 There is little information available about the interaction between these two bacterial pathogens and the importance of this interaction with regards to the severity and outcome of infection. Some studies that have examined the interaction of P. aeruginosa and S. aureus suggest induction of adaptive changes in S. aureus which could promote its virulence. For example, P. aeruginosa could induce S. aureus to mount an adaptive response by forming small-colony variants.Citation25,Citation27,Citation28 It has also been demonstrated that P. aeruginosa-produced quinolone 4-hydroxy-2-heptylquinoline-N-oxide (HQNO) could suppress growth of S. aureus and a wide spectrum of other Gram-positive bacteria.Citation29,Citation30 The same molecule can also induce resistance to streptomycin or tobramycin.Citation25,Citation31 In wounds with complex infections, pathogenicity of S. aureus has been shown to increase in the presence of low concentrations of P. aeruginosa.Citation32
We isolated two pigment variants of S. aureus and one strain of P. aeruginosa from the same clinical sample obtained from a patient with a soft tissue wound. S. aureus pigment variants produced white and yellow colonies. In this study we report that the white colony variant harbors an intact crtOPQMN operon in spite of lack of pigmentation. Furthermore, we demonstrate that the crtOPQMN operon of the white variant is functional and can be triggered to produce golden pigment by the P. aeruginosa isolate. We also show that catalase, another S. aureus virulence factor, is also upregulated as a consequence of its interaction with the P. aeruginosa. The phenotypic changes induced by the co-infecting P. aeruginosa correlates to increased resistance of the white S. aureus variant to stressful conditions.
Materials and methods
Identification of bacteria
The two phenotypic variants of S. aureus and the P. aeruginosa strain were initially identified by an automated system for identification of bacteria (Phoenix Automated Microbiology System, Sparks, MD, USA) according to the manufacturer instructions. These results were confirmed by 16 s ribosomal DNA sequencing. Briefly, after isolation of genomic materials from both phenotypic variants, eubacterial universal 16S rDNA primers, 27 A and 1392RCitation33 (), were used to amplify the entire 16S rDNA gene. The PCR amplicons were cloned into a TA-cloning vector, clonally expanded by transformation into E. coli, and sequenced by the dideoxy Sanger method. The three isolates were designated as S. aureus white and S. aureus yellow variants, and P. aeruginosa 6611.
Table 1 Sequencing and qRT-PCR primers
Sequencing of crtOPQMN operon
Known sequences of crtOPQMN operon deposited in the GenBank were used in multiple sequence alignment to identify conserved regions for the design of PCR primers. These primers () were then used for the amplification of overlapping segments covering the entire operon, including the promoter region. The amplicons were either directly sequenced or cloned into a TA-vector before sequencing. The sequencing data from the amplicons were used to assemble the entire operon including its promoter region. All sequence manipulation was carried out using commercially available software (v5.4.5; Geneious Pro, Biomatters Ltd, Newark, NJ, USA).
Co-culture experiment for induction of pigment production
Bacteria used in these experiments were grown in Luria-Bertani (LB) broth at 37°C, 220 rpm for 18–24 hours. P. aeruginosa 6611 was grown in stationary conditions in LB broth as a biofilm at 37°C. For co-culture experiments, an overnight culture of S. aureus white variant was used. P. aeruginosa 6611 for co-culture experiments was prepared from a 72-hours-old biofilm. Biofilm was disrupted by pipetting and vigorous vortexing. Cells were pelleted by spinning at 13,000 g for 5 minutes, supernatant was decanted, and the pellet was re-suspended in fresh LB broth and used in co-culture experiments.
Co-culture experiments were carried out by inoculating 2 μL of P. aeruginosa 6611 near 5 μL of of S. aureus white variant on semi-solid LB agar plates such that the distance between the points of inoculations were a constant 1 cm. Agar plates were incubated upside down at 37°C and monitored for pigment development for 48 hours. As the experimental control, the white variant was also similarly co-cultured with Escherichia coli Mg1655, Acinetobacter baumani, co 41-00-sc1-c1, Enterobacter W001, and the white and yellow variants of S. aureus.
We also examined the supernatants from the biofilm and shaking cultures of P. aeruginosa for their ability to induce pigment production in the white S. aureus variant. The supernatants were sterile filtered through a 0.22 μm filter and spotted on an LB agar plate near the white variant. However, we did not observe any pigment induction using this method, even though the supernatant was inoculated multiple times at the same spot during the experiment.
Quantitative real-time polymerase chain reaction (qRT-PCR) of genes involved in staphyloxanthin synthesis and control of its expression
To prepare RNA for qRT-PCR, co-culture experiments were carried out by incubation of 5 μL of S. aureus white variant overnight culture on LB agar plates (six spots per plate) for 18–24 hours. P. aeruginosa 6611 was then spotted at 1 cm distance (see above) from the center of the S. aureus colonies and returned to the incubator. A plate containing only white colonies was also prepared as a control. At different time points of 1, 7, 24, and 48 hours from the start of the co-cultures, S. aureus colonies were collected and re-suspended in RNAprotect Bacteria Reagent (Qiagen, Valencia, CA, USA) according to the manufacturer instructions and preserved at −80°C until RNA extraction.
RNA was extracted using the Ribo-Pure RNA extraction kit (Ambion®, Grand Island, NY, USA). The quantity and quality of RNA was evaluated using a NanoDrop1000 spectrophotometer and Agilent Bioanalyzer 2100, respectively. The genes examined in this study were crtM, sigma B transcription factor (sigB), global transcriptional regulator (sarA), FPP synthase gene, post-transcriptional regulatory protein Hfq gene (hfq), accessory gene regulator (agrA), and another important virulence factor for H2O2 survival, catalase (katA). The sequence of crtOPQMN from the white variant was used to design qRT-PCR primers for crtM gene. We used the 16S rRNA gene as the internal control for the normalization of gene expression. Primers for other selected genes were designed based on the consensus sequence constructed from their known sequences deposited in the GenBank. Quantitative RT-PCR was performed using the Applied Biosystems 7500 and Power SYBR Green RNA to Ct 1-step kit (Applied Biosystems, Carlsbad, CA, USA) according to manufacturer instructions using 20 ng of total RNA per reaction. The values of Ct were calculated by 21 CFR Part 11 SDS software (v 1.4.1; Applied Biosystems, Carlsbad, CA, USA) with the baseline set from 3–6 cycles, and a threshold value of 0.2. Data were analyzed using the ∆∆Ct method.Citation34 The results represent the average fold change of two independent experiments comparing the white variant with or without co-culturing with P. aeruginosa 6611.
Staphyloxanthin analysis
For spectroscopic analysis, staphyloxanthin was extracted as previously described.Citation35 Briefly, the S. aureus white variant was grown in the presence (co-culture) and absence of P. aeruginosa 6611 for 48 hours. The pigmented (yellow) S. aureus variant was also cultured on plates in the absence of P. aeruginosa 6611, and used as a positive control. After incubation, bacteria were collected from the LB agar plates and re-suspended in sterile Dulbecco’s phosphate-buffered saline (DPBS). Optical density (OD600) of the suspensions was adjusted to 0.50 ± 0.05, and 5 mL of the OD-adjusted suspensions was used for the pigment extraction. The cells were collected by centrifugation for 5 minutes at 13,000 g, re-suspended in 200 μL of 200 proof methanol (Sigma-Aldrich, St Louis, MO, USA), and heated for 5 minutes at 55°C. The cells were pelleted by centrifugation for 1 minute at 13,000 g and the supernatants were collected. The extraction step was repeated once more. Methanol extracts from both rounds of extractions were pulled, their volumes were adjusted to 1 mL, and the absorbance of the crude extract was measured at λ = 465 nm using a Genesys 20 spectrophotometer (Thermo Electron Corporation, East Greenbush, NY, USA).
For high performance thin layer liquid chromatography (HPTLC), methanol extracts were protected from light and vacuum evaporated at 45°C. After drying, samples were dissolved in 10 μL of ethyl acetate (Sigma-Aldrich) and spotted on the HPTLC silica plates with a pore size of 60 Å (Sigma-Aldrich). The plates were placed in a TLC tank containing 65 mL chloroform (Sigma-Aldrich), 24 mL methanol (Sigma-Aldrich), and 4 mL deionized water.Citation36 The spots were allowed to migrate upward for 15 minutes, and the plate was then air-dried and photographed.
Survival in H2O2
After co-culturing of the S. aureus white variant with P. aeruginosa 6611 for 48 hours, pigment-induced colonies were collected from the LB agar and re-suspended in sterile DPBS. The white variant incubated in the absence of P. aeruginosa 6611 was used as the control in these experiments. The optical densities of the bacterial suspensions were adjusted to OD600 = 0.15 in DPBS. The number of bacteria was then adjusted to 20,000 colony forming units (CFU) per mL by serial dilutions in DPBS. Five hundred microliters of bacterial suspension containing 10,000 CFU was mixed with a final concentration of 1% H2O2 in 14 mL polystyrene tubes. Cells were incubated for 15 minutes at 37°C with shaking at 220 rpm. After 15 minutes of incubation with H2O2, catalase at a final concentration of 1000 U/mL was added, and incubated for 15 minutes to stop the reaction. Finally, cells were plated on LB agar with appropriate dilutions and CFUs were determined after 18–24 hours of incubation at 37°C. Survival rates were expressed as a percentage of the initial number of cells. Results are presented as the mean value of two independent experiments.
Survival in 1 mM polymyxin B
The S. aureus white variant with or without co-culturing with P. aeruginosa 6611 was collected from LB agar plates and the number of cells adjusted to 104 CFU/mL in sterile DPBS. Polymyxin B (Sigma-Aldrich) at a final concentration of 1 mM was added to each sample and incubated for 30 minutes at 37°C in 3 mL polystyrene tubes. After polymyxin B treatment, bacteria were appropriately diluted and plated on LB agar, and the number of CFUs was determined after 18–24 hours. The survival rates were expressed as the percentage of the initial number of cells, and presented as an average of three independent experiments.
Intracellular survival of S. aureus in mouse macrophage cell line
Mouse macrophage cell line (J7741.A, ATCC, Manassas, VA, USA) was cultured according to the manufacturer instructions with ATCC-formulated Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 500 U/mL penicillin/streptomycin solution. Cells were maintained and subcultured at 37°C in an incubator in the presence of 5% CO2. For in vitro infection experiments, cells were seeded in 6-well plates and grown until a monolayer was formed (~1.7 × 106 cells/monolayer). Approximately 24 hours before the addition of bacteria, growth media containing penicillin/streptomycin was replaced with invasion media (DMEM and 10% FBS without antibiotics). For the infection assay, 1.7 × 107 bacteria was added to each well (infection ratio of 1:10) and incubated at 37°C. After 30 minutes of incubation, extracellular bacteria were removed and macrophage monolayer was rinsed with 1 mL of DPBS. The infected monolayer was then incubated with 2 mL of fresh media containing 100 μg/mL of gentamicin. At different time intervals of 15 minutes (T0) and 120 minutes (T1) after the addition of the bacteria, the gentamicin-containing medium was removed cells were washed once with DPBS, and lysed with 0.02% Triton X-100 in DPBS. Appropriate dilutions were then plated on LB agar and the number of CFUs determined after 18–24 hours of incubation at 37°C. Intracellular survival was expressed as a percentage of CFUs comparing the number of cells at T0 and T1. The survival rates were presented as the mean of three independent experiments.
Statistical analysis
Results are presented as mean ± SD. Repeats of three independent experiments were used for statistical analysis which was performed by JMP10® software (SAS Institute Inc, Duke University, Cary, NC, USA). Student’s t-test was used for the evaluation of the statistical significance between the groups. P-values < 0.05 were considered to be significant.
Results
Sequencing of crtOPQMN operon and its promoter region
We initially postulated that the difference between the two pigment variants of S. aureus was due to the differences in sequences of the coding or promoter regions of the crtOPQMN. Eight primer pairs were designed and used to amplify overlapping segments of the crtOPQMN operons generating amplicons ranging in size from 865 to 1017 bps (). The amplicons were sequenced and assembled to construct the full length of the operon from both the white and yellow variants of S. aureus. Comparison of these two operons, including the promoter regions, indicated that they were identical. Both operons contained the same gene arrangement as the canonical crtOPQMN operon: the promoter followed by genes coding for glycosyl-4,4′-diaponeurosporenoate acyltransferase (crtO), phytoene dehydrogenase (crtI), 4,4′-diaponeurosporenoate glycosyltransferase (crtQ), dehydrosqualene synthase (crtM), and dehydrosqualene desaturase (crtN). This indicated that some mechanism other than mutation in the crtOPQMN genes or the promoter was responsible for the lack of pigment production in the white variant.
Figure 1 Positions of overlapping primers on the crtOPQMN operon.
Note: The green triangles indicate the position of overlapping primers used for the amplification and sequencing of the crtOPQMN operon.
Abbreviations: crtOPQMN, carotenoid biosynthetic operon; F, forward; R, reverse; crtO, glycosyl-4,4′-diaponeurosporenoate acyltransferase; crtI, phytoene dehydrogenase; crtQ, 4,4′-diaponeurosporenoate glycosyltransferase; crtM, dehydrosqualene synthase; crtN, dehydrosqualene desaturase.

Induction of pigment production in white phenotypic variant and pigment analysis
In some bacteria, pigment production is controlled by quorum-sensing signals.Citation37–Citation39 It has also been demonstrated that P. aeruginosa affects gene expression in S. aureus through secreted quorum-signaling molecules.Citation28 We sought to determine if P. aeruginosa 6611 isolated along with the white S. aureus variant could induce pigment production in the latter. Interestingly, after 24 hours of co-culturing with P. aeruginosa 6611 the white S. aureus variant developed golden pigment, the intensity of which maximized at 48 hours. This induction of pigment was not seen when the white variant was co-cultured with bacterial strains Escherichia coli Mg1655, Acinetobacter baumani co 41-00-sc1-c1, and Enterobacter W001 (). To quantify pigment production in the white variant, we carried out a co-culture experiment between P. aeruginosa 6611 and the white variant. We also used the pigmented yellow variant in the co-culture experiments as a positive control. Spectrophotometric measurement of absorbance (A465) of crude methanol extracts of the white variant at 48 hours showed an approximately 4-fold increase in absorbance. We did not observe any such increase of absorption in crude extracts from the yellow variant. Interestingly, the absorbance (A465) of crude extracts from S. aureus white and yellow variants, after co-culture with P. aeruginosa 6611, were comparable (data not shown). This observation suggests that co-culture with P. aeruginosa had minimal effect on the yellow variant’s pigment production. The extracted pigments were also analyzed by HPTLC, which clearly demonstrated the induction of pigment production in the white phenotypic variant after co-culture with P. aeruginosa 6611 (). Therefore, the crtOPQMN operon in the white variant of S. aureus is functional and staphyloxanthin production can be induced by environmental factors such as secreted molecules from P. aeruginosa 6611.
Figure 2 The effect of co-culture with different bacteria on pigment production by the Staphylococcus aureus white variant.
Notes: The co-culture experiment was carried out on an LB agar plate for 48 hours between the Staphylococcus aureus white variant (W) and Escherichia coli Mg1655, Acinetobacter baumani/co 41-00-sc1-c1, Enterobacter W001, or Pseudomonas aeruginosa 6611. Only P. aeruginosa 6611 induced pigment production in the white variant (yellow colony).
Abbreviation: LB, luria-Bertani.
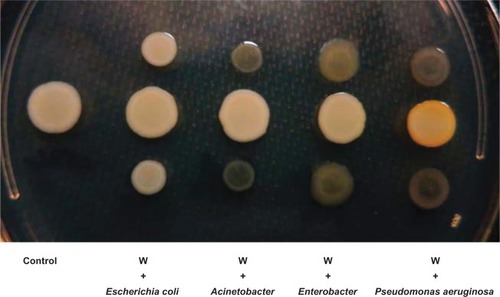
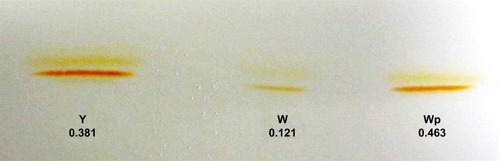
Figure 3 HPTLC analysis of staphyloxanthin in crude methanol extract from Staphylococcus aureus. Crude methanol extracts of staphyloxanthin from the S. aureus white variant (W), the S. aureus white variant co-cultured for 48 hours with Pseudomonas aeruginosa 6611 (Wp), and the S. aureus yellow variant (Y) were analyzed by HPTLC on silica plates (pore size 60 Å). The results indicated a significant difference in pigment-band intensity between the white variant and the white variant co-cultured with P. aeruginosa 6611.
Notes: Numbers below the bands represent the absorbance of the crude methanol extracts at A465 (W = 0.121, Wp = 0.463, Y = 0.381). There is ~4-fold increase in absorbance in crude extracts of the white variant after co-culture with P. aeruginosa 6611, indicating substantial increase in pigment production.
Abbreviation: HPTLC, high performance thin layer liquid chromatography.
Quantitative RT-PCR of genes relevant for expression and/or control of staphyloxanthin
Since the crtOPQMN operon of the white variant has an intact coding region and promoter, the induction of staphyloxanthin production by P. aeruginosa 6611 is either controlled at a step upstream of the staphyloxanthin biosynthetic pathway, or at a post-transcriptional step. We examined the expression of the crtM gene as well as several other genes that could control the crtOPQMN operon.
The expression of crtM did not change significantly after 1 hour and 7 hours of co-culture with P. aeruginosa 6611 (). Although there was a slight increase in expression at 7 hours (~1.7-fold), this may not represent a significant change. The expression of genes coding for transcription factor sigma B (sigB) and global transcriptional regulator (sarA), both of which have been shown to positively control crtOPQMN operonCitation10,Citation40 also did not change significantly after the first and seventh hours. The gene for farnesyl diphosphate synthase (FPP synthase gene) is involved in the synthesis of farnesyl diphosphate, which is the substrate for the first step of the staphyloxanthin biosynthetic pathway. Expression of this gene also did not change during the early time intervals. Post-transcriptional regulator gene (hfq) has recently been shown to participate in RNA-mediated inhibition of crtOPQMN operon translation.Citation41 The expression of this gene was also not affected during the first two time intervals. With the exception of sarA, expression of the abovementioned genes was significantly downregulated after a 24-hour co-culture with P. aeruginosa 6611.
Figure 4 Transcriptional analysis of selected genes from the Staphylococcus aureus white variant after co-culture with Pseudomonas aeruginosa. Several genes involved in pigment production were analyzed by qRT-PCR after co-culture with P. aeruginosa 6611. These genes included crtM, which is directly involved in staphyloxanthin synthesis. Transcriptional factors SigB and Sara control the expression of the crtOPQMN operon, and product of the hfq gene may be involved in post-transcriptional control of the operon. FPP-synthase gene is involved in the synthesis of farnesyl-diphosphate which is the substrate for the crtOPQMN operon. The results indicate no significant changes in expression of these genes through the first 7 hours of co-culture. In the later time intervals of 24 hours and 48 hours, these genes (with the exception of sarA which was slightly downregulated) were significantly downregulated. The agrA gene could be involved in the downregulation of staphyloxanthin. The expression of the katA gene may be correlated to the production of staphyloxanthin since both confer resistance to H2O2. The transcription of agrA is downregulated after 7 hours and into the later time intervals. In contrast, katA gene is significantly upregulated after 8 hours and reached its peak (more than 3-fold) at 24 hours.
Notes: The columns represent fold change in gene expression of co-cultured S. aureus white variant as compared to the white variant that was not co-cultured. Time points represent data gathered after 1, 7, 24, and 48 hours after the start of co-culture with P. aeruginosa 6611.
Abbreviations: qRT-PCR, quantitative reverse transcription polymerase chain reaction; crtM, carotenoid biosynthetic gene M; sigB, sigma B transcription factor gene; sarA, staphylococcal accessory regulator A gene; FPP synth, farnesyl pyrophosphate synthase gene; hfq, host factor gene for Q beta; katA, catalase gene A; agrA, accessory gene regulator A; crtOPQMN, carotenoid biosynthetic operon.
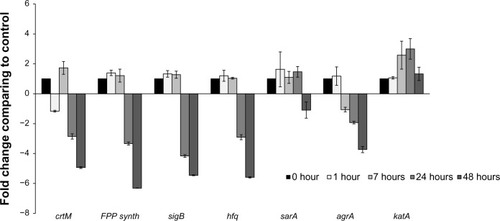
We also examined the expression of two other genes that code for two important virulence factors, AgrA and catalase. A component of the AgrC/AgrA two-component signal transduction system, increased expression of Staphylococcal accessory regulator gene (agrA) promotes dispersal of S. aureus biofilms,Citation42 and downregulates staphyloxanthin biosynthetic genes.Citation43 Catalase is an important virulence factor which protects S. aureus against H2O2.Citation44 The expression of catalase could be coordinated with the expression of staphyloxanthin to provide the maximum protection against H2O2. Therefore, we also examined the expression of catalase in co-culture experiments. The expression of agrA followed a pattern similar to other genes examined and was down-regulated after 7 hours. In contrast, catalase transcription was upregulated to about 2.5-fold after 7 hours and more than 3-fold after 24 hours. Both downregulation of agrA and upregulation of catalase can be considered to promote the survival of S. aureus in stressful conditions. The trend in expression of genes examined in this study could represent a global adaptive response by S. aureus to the presence of P. aeruginosa 6611, which consists of slowing of metabolism, promotion of biofilm formation, and increased resistance to oxygen free radicals.
Survival in the presence of H2O2
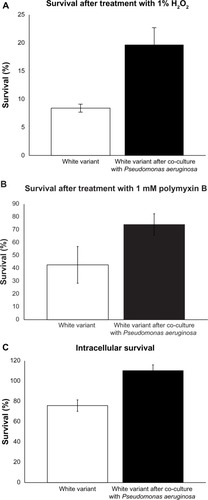
Staphyloxanthin, due to its antioxidant properties, has a role in survival in hyperoxic environments and during neutrophilic bursts.Citation13 We sought to determine if pigment induction in the S. aureus white variant can enhance survival in the presence of H2O2. After co-culturing the white variant with P. aeruginosa 6611, the newly pigmented white variants were treated with 1% H2O2 and accessed for their survival as compared to the non-pigmented white variants. Our results indicated that survival of the white variant significantly improved from 8.9% ± 0.7% in absence of pigmentation to 21.8% ± 3.1% after P. aeruginosa-induced pigment production (). The role of staphyloxanthin in the protection of S. aureus against H2O2 is well established by previous independent studies,Citation12,Citation14 and our experiments served merely as confirmation of those results. Therefore, we believe our results are valid in demonstrating the importance of staphyloxanthin in the survival of bacterium in a hyperoxic environment despite only two repeats of independent experiments.
Figure 5 Induction of resistance to stress in the Staphylococcus aureus white variant by Pseudomonas aeruginosa 6611. (A) co-culture with P. aeruginosa 6611 for 48 hours led to enhanced survival of the white variant in the presence of 1% H2O2 as compared to the white variant without co-culturing (21.8% ± 3.1% vs 8.9% ± 0.7%, P < 0.05). Induction of staphyloxanthin production and upregulation of catalase gene could account for increased resistance of the white variant to H2O2. (B) co-culture with P. aeruginosa 6611 for 48 hours increased the resistance of the white variant to 1 mM polymyxin B. The difference between survival of the white variant after co-culturing and the white variant without co-culturing is almost 2-fold (75.3% ± 8.3% vs 42.7% ± 14.9%). (C) co-culture with P. aeruginosa 6611 for 48 hours resulted in enhanced survival of the white variant within the mouse macrophage cell line J7741.a. after 2 hours of infection, the number of co-cultured white variant increased slightly (110.4% ± 4.1%). In the parallel experiment with the white variant without co-culturing, the number of survived bacteria decreased after 2 hours of infection (75.9% ± 3.9%, P < 0.05).
Notes: All results represent the mean from either two experiments (panel A) or three experiments (panels B and C), expressed as a percentage of the initially inoculated number of the S. aureus white variant (white bars) and the pigmented white variant after 48 hours of co-culture with P. aeruginosa 6611 (black bars). The P-value was determined using student’s t-test.
Survival in the presence of polymyxin B
Polymyxin B is a broad spectrum antibiotic that is mostly used against Gram-negative bacteria, but has also been shown to be effective against S. aureus.Citation45 Polymyxin B binds to the lipopolysaccharide layer of the bacterial cell wall with its fatty acid portion inserted into the hydrophobic region of the plasma membrane. This increases membrane fluidity, allowing water to enter the cytoplasm. We hypothesized that staphyloxanthin production, due to its membrane stabilization effects, will increase bacterial survival during exposure to polymyxin B. As expected, the induction of staphyloxanthin production after co-culture with P. aeruginosa 6611 increased resistance of the white variant to this antibiotic. Pigmented S. aureus white variants had a survival rate of 75.3% ± 8.3%, as compared to 42.7% ± 14.9% for the non-pigmented white variants (). Despite the almost two times higher survival rate of the pigment-induced and non-pigmented white variants, this difference failed to reach threshold for statistical difference. However, this data supports the view of staphyloxanthin stabilizing activity in the plasma membrane.
Intracellular survival
We examined the impact of P. aeruginosa 6611 co-culture on the white variant’s survival within macrophages. The mouse macrophage cell line J7741.A was infected for 2 hours with P. aeruginosa-induced pigmented white variant and its survival was compared to the non-pigmented white variant. We calculated the intracellular survival rate as the percentage of the initial (T0) number of bacteria that survived after 2 hours (T1). Our results showed 110.4% ± 4% intracellular survival of pigmented bacteria as compared to 75.9% ± 3.9% for the non-pigmented bacteria (P < 0.05) (). The 110% represents an approximately 10% increase in the population of pigmented white variant after 2 hours within the intracellular environment of macrophages. These results suggest that S. aureus white variant co-cultured with P. aeruginosa 6611, not only survived better than the control white variant, but may have also divided within the macrophages during the 2-hour infection period. The data from this experiment indicates a clear intracellular survival advantage for S. aureus white variant after co-culture with P. aeruginosa 6611.
Discussion
Interspecies interactions play a significant role in wound infections since these infections are commonly polymicrobial with constant physical and chemical communication between the microbes. Modulation of gene expression as a result of this interaction could have great impact on the pathogenicity of microbes and consequently clinical outcomes of the infection. This study demonstrates the importance of such interspecies communication with respect to pathogenic phenotypes of coinfecting bacteria. P. aeruginosa, which is often isolated along with S. aureus from wound infections,Citation46 induced the production of at least two virulence factors, staphyloxanthin and catalase, in a white variant of S. aureus. This had a direct effect on its resistance to stress and survival within host macrophages. Golden pigment staphyloxanthin and catalase enzyme are both antioxidative defense mechanisms that confer survival advantage during infection. Staphyloxanthin also provides protection against membrane-disrupting antimicrobial peptides.
An important finding in this study was that a naturally occurring white variant of S. aureus carries an intact and functional crtOPQMN operon for the synthesis of golden pigment staphyloxanthin. The fact that this operon does not produce staphyloxanthin in the absence of environmental stimuli (eg, the molecules originated from P. aeruginosa) indicates an intricate regulatory mechanism in the control of pigment production. More interesting may be the mechanism of this control system. We examined the transcription of several genes that are directly or indirectly involved in the synthesis of staphyloxanthin. These included the crtM gene that mediates the first reaction in the staphyloxanthin biosynthetic pathway, as well as transcription factors sigB and sarA genes which positively control its expression. P. aeruginosa-induction of pigment production in the S. aureus white variant did not lead to upregulation in the transcription of crtM or the two transcription factors. This finding is contrary to previous studies that reported increased transcription of sigB after exposure to signals from P. aeruginosa.Citation28 This discrepancy between our results and those of others may be partly due to distinct bacterial strains used in different studies. Furthermore, in choosing time points of 1, 7, 24, and 48 hours after the addition of P. aeruginosa 6611 in co-culture experiments, we may have missed transient upregulation of genes involved in staphyloxanthin biosynthesis.
We also explored the possibility that synthesis of the upstream substrate for staphyloxanthin synthesis could be affected by P. aeruginosa. The substrate of the first enzymatic reaction for S. aureus staphyloxanthin biosynthesis is (2E,6E)-farnesyl diphosphate, which itself is synthesized from isopentenyl diphosphate through a 3-step enzymatic process. Farnesyl-diphosphate synthase, which also has geranyltranstransferase activity, carries out the last two steps of this process. Examination of farnesyl-diphosphate synthase mRNA transcript indicated no upregulation after 7 hours of incubation of the S. aureus white variant with P. aeruginosa 6611. Furthermore, after 24 hours, transcription of the gene coding for this enzyme was significantly decreased, as was the case with most other genes analyzed in this study.
Control and modulation of gene expression can also be accomplished by post-transcriptional mechanisms. In E. coli, the Hfq protein mediates interaction between some small RNA species and their target mRNAs, which could affect mRNA translation. In a recent study, Liu et al showed that deletion of the hfq gene in a strain of S. aureus led to increased expression of surface pigment.Citation47 It appears that in certain strains of S. aureus, Hfq may directly interact with the target mRNA, thereby controlling gene expression at the level of translation. We did not see any significant decrease in transcription of the hfq gene in the white S. aureus variant after 7 hours of incubation with P. aeruginosa 6611. However, at the 24- and 48-hour time intervals, there was considerable decrease in transcription of most of the genes examined, including the hfq gene. Since P. aeruginosa-induced pigmentation appears after 24 hours, we cannot rule out a role for Hfq in mediating this outcome. Therefore, the exact mechanism by which P. aeruginosa 6611 controls staphyloxanthin production in the white S. aureus variant remains to be elucidated.
We also examined the expression of AgrA, (a part of the AgrC/AgrA two-component signal transduction system) which positively regulates the dispersal of S. aureus biofilmsCitation42 and downregulates the expression of staphyloxanthin.Citation44 Expression of the agrA gene did not change during the first 7 hours of co-culture with P. aeruginosa 6611. The expression of agrA was, however, significantly downregulated after 24 hours and correlated to upregulation of pigment production. Therefore, downregulation of the agrA gene in presence of P. aeruginosa 6611, may promote pigment production in the white variant.
Catalase gene, is considered an important virulence factor which protects S. aureus against H2O2.Citation43 Since both catalase and staphyloxanthin protect the microorganisms against the same stress condition, it is plausible that their expression would be coordinated. Interestingly, the transcription of the catalase gene of the white S. aureus variant was significantly upregulated after 7 hours of co-culture with P. aeruginosa 6611 and peaked after 24 hours. Although the transcription of the catalase gene contrasts that of other genes, it is consistent with the trend in the adaptation of the white variant to a more H2O2-resistant phenotype. This bacterium appears to responds to P. aeruginosa 6611 stimulation by changing its gene expression to acquire a more stress-resistant phenotype, enabling it to withstand oxidative stress, as well as the intracellular environment of macrophages.
Although our results demonstrating the antioxidant role of staphyloxanthin is consistent with those of other authors, it is important to draw a clear distinction between these studies. In other studies, gene knockout experimentsCitation13 and antagonist drugs targeting crtOPQMN operonCitation17,Citation36,Citation48 were used to show the importance of staphyloxanthin for the survival of S. aureus in hyperoxidative environments. In our study, the S. aureus white variant was a naturally occurring bacterium isolated from a clinical sample, rather than being artificially constructed by genetic manipulation or by the administration of antagonistic drugs. Furthermore, our study demonstrates an important example of interspecies interaction that may be occurring in polymicrobial infections.
The medical community is beginning to recognize the significance of polymicrobial infections and the role of the human-associated microbial community in promoting or averting such infections. Many therapies are now taking into account the polymicrobial etiology of diseases and the attendant implications for treatment and prevention.Citation23 Our results demonstrate that P. aeruginosa 6611 can induce transcriptional changes in S. aureus, which contribute to its virulence and intracellular survival. We show that a phenotypically white variant of S. aureus carries a functional apparatus for pigment synthesis, which can be activated by a co-infecting P. aeruginosa 6611, presumably via secreted signals. Additionally, another S. aureus virulence factor, catalase, can also be induced as a result of this interspecies interaction. To the best of our knowledge, this is the only example in the literature thus far demonstrating interaction between co-infecting S. aureus and P. aeruginosa which could impact the virulence and intracellular survival of at least one of them. Further investigation of interactions between S. aureus and P. aeruginosa will provide deeper understanding of the mechanisms that control their virulence.
Acknowledgments
The authors wish to thank the Combat Wound Initiative Program, and the Henry M Jackson Foundation for the Advancement of Military Medicine. The views expressed in this manuscript are those of the authors and do not reflect the official policy of the Department of the Army, the Department of Defense, or the US Government. This effort was supported, in part, by the Combat Wound Initiative Program. Some of the authors are military service members. This work was prepared as part of their official duties. We certify: (1) that all individuals who qualify as authors have been listed; (2) that each has participated in the conception and design of this work, the analysis of data (when applicable), the writing of the document, and/or the approval of the submission of this version; (3) that the document represents valid work; (4) that if we used information derived from another source, we obtained all necessary approvals to use it and made appropriate acknowledgments in the document; and, (5) that each author takes public responsibility for it.
Disclosure
The authors report no conflicts of interest in this work.
References
- LennetteEHBalowsAHauslerWJJrShadomyJHManual of Clinical Microbiology4th edWashington, DCAmerican Society for Microbiology1985
- RosenbachAJMikro-Qrganismen bei den Wund Infections Krankheiten des MenschenWiesbadenJF Bergmann1884 German
- RosenfeldMRamseyBWGibsonRLPseudomonas acquisition in young patients with cystic fibrosis: pathophysiology, diagnosis, and managementCurr Opin Pulm Med20039649249714534401
- McDougallKEPerryMJGibsonRLColleySMKorachKSTobiasJHEstrogen receptor-alpha dependency of estrogen’s stimulatory action on cancellous bone formation in male miceEndocrinology200314451994199912697707
- BartowskyEJXiaDGibsonRLFleetGHHenschkePASpoilage of bottled red wine by acetic acid bacteriaLett Appl Microbiol200336530731412680944
- MashburnKLAtkinsonSEvaluation of adrenal function in serum and feces of Steller sea lions (Eumetopias jubatus): influences of molt, gender, sample storage, and age on glucocorticoid metabolismGen Comp Endocrinol2004136337138115081837
- HoltDCHoldenMTTongSYA very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthinGenome Biol Evol2011388189521813488
- MarshallJHWilmothGJProposed pathway of triterpenoid carotenoid biosynthesis in Staphylococcus aureus: evidence from a study of mutantsJ Bacteriol198114739149197275937
- MarshallJHWilmothGJPigments of Staphylococcus aureus, a series of triterpenoid carotenoidsJ Bacteriol198114739009137275936
- PelzAWielandKPPutzbachKHentschelPAlbertKGotzFStructure and biosynthesis of staphyloxanthin from Staphylococcus aureusJ Biol Chem200528037324933249816020541
- WielandBFeilCGloria-MaerckerEGenetic and biochemical analyses of the biosynthesis of the yellow carotenoid 4,4′-diaponeurosporene of Staphylococcus aureusJ Bacteriol199417624771977268002598
- ClauditzAReschAWielandKPPeschelAGotzFStaphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stressInfect Immun20067484950495316861688
- LiuGYEssexABuchananJTStaphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activityJ Exp Med2005202220921516009720
- Popov IuAKaprel’iantsASOstrovskiiDNIgnatovVVStudy of the membranes of pigment-free mutant of Staphylococcus aureusBiokhimiia197641611161120 Russian1027489
- MishraNNLiuGYYeamanMRCarotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptidesAntimicrob Agents Chemother201155252653121115796
- LiuCILiuGYSongYA cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulenceScience200831958681391139418276850
- SongYLiuCILinFYInhibition of staphyloxanthin virulence factor biosynthesis in Staphylococcus aureus: in vitro, in vivo, and crystallographic resultsJ Med Chem200952133869388019456099
- ValenciaICKirsnerRSKerdelFAMicrobiologic evaluation of skin wounds: alarming trend toward antibiotic resistance in an inpatient dermatology service during a 10-year periodJ Am Acad Dermatol200450684584915153882
- BrookIFrazierEHAerobic and anaerobic microbiology of chronic venous ulcersInt J Dermatol19983764264289646126
- MousaHAAerobic, anaerobic and fungal burn wound infectionsJ Hosp Infect19973743173239457609
- SummanenPHTalanDAStrongCBacteriology of skin and soft-tissue infections: comparison of infections in intravenous drug users and individuals with no history of intravenous drug useClin Infect Dis199520Suppl 2S279S2827548575
- BowlerPGDuerdenBIArmstrongDGWound microbiology and associated approaches to wound managementClin Microbiol Rev200114224426911292638
- BrogdenKAGuthmillerJMTaylorCEHuman polymicrobial infectionsLancet2005365945525325515652608
- HarrisonFMicrobial ecology of the cystic fibrosis lungMicrobiology2007153Pt 491792317379702
- HoffmanLRDezielED’ArgenioDASelection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosaProc Natl Acad Sci U S A200610352198901989517172450
- KorgaonkarATrivediURumbaughKPWhiteleyMCommunity surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infectionProc Natl Acad Sci U S A201311031059106423277552
- BiswasLBiswasRSchlagMBertramRGotzFSmall-colony variant selection as a survival strategy for Staphylococcus aureus in the presence of Pseudomonas aeruginosaAppl Environ Microbiol200975216910691219717621
- MitchellGSeguinDLAsselinAEStaphylococcus aureus sigma B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2-heptylquinoline-N-oxideBMC Microbiology2010103320113519
- LightbownJWJacksonFLInhibition of cytochrome systems of heart muscle and certain bacteria by the antagonists of dihydrostreptomycin: 2-alkyl-4-hydroxyquinoline N-oxidesBiochem J195663113013713315258
- MachanZAPittTLWhiteWInteraction between Pseudomonas aeruginosa and Staphylococcus aureus: description of an anti-staphylococcal substanceJ Med Microbiol19913442132171902262
- LightbownJWJacksonFLInhibition of cytochrome system of heart muscle and of Staphylococcus aureus by 2-heptyl-4-hydroxyquin-oline-N-oxide, an antagonist of dihydrostreptomycinBiochem J1954584xlix13230048
- HendricksKJBurdTAAnglenJOSimpsonAWChristensenGDGainorBJSynergy between Staphylococcus aureus and Pseudomonas aeruginosa in a rat model of complex orthopaedic woundsJ Bone Joint Surg Am200183-A685586111407793
- MarchesiJRSatoTWeightmanAJDesign and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNAAppl Environ Microbiol19986427957999464425
- PfafflMWA new mathematical model for relative quantification in real-time RT-PCRNucleic Acids Res2001299e4511328886
- MorikawaKMaruyamaAInoseYHigashideMHayashiHOhtaTOverexpression of sigma factor, sigma(B), urges Staphylococcus aureus to thicken the cell wall and to resist beta-lactamsBiochem Biophys Res Commun2001288238538911606054
- SongYLinFYYinFPhosphonosulfonates are potent, selective inhibitors of dehydrosqualene synthase and staphyloxanthin biosynthesis in Staphylococcus aureusJ Med Chem200952497698819191557
- WilliamsonNRFineranPCLeeperFJSalmondGPThe biosynthesis and regulation of bacterial prodigininesNat Rev Microbiol200641288789917109029
- McCleanKHWinsonMKFishLQuorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactonesMicrobiology1997143Pt 12370337119421896
- ThomsonNRCrowMAMcGowanSJCoxASalmondGPBiosynthesis of carbapenem antibiotic and prodigiosin pigment in Serratia is under quorum sensing controlMol Microbiol200036353955610844645
- FeyPDEndresJLYajjalaVKA genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genesMBio20134e005371223404398
- CastroSLNelman-GonzalezMNickersonCAOttCMInduction of attachment-independent biofilm formation and repression of Hfq expression by low-fluid-shear culture of Staphylococcus aureusAppl Environ Microbiol2011776368637821803898
- AbdelnourAArvidsonSBremellTRydenCTarkowskiAThe accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis modelInfect Immun1993619387938858359909
- QueckSYJameson-LeeMVillaruzAERNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureusMol Cell20083215015818851841
- KanafaniHMartinSECatalase and superoxide dismutase activities in virulent and nonvirulent Staphylococcus aureus isolatesJ Clin Microbiol1985216076103988902
- DeguchiKYokotaNKoguchiMAntimicrobial activities of polymyxin B against clinically isolated microbial strains. Results of MIC determination including high concentrationsJpn J Antibiot19934610884895 Japanese8254890
- VyasRGaoGYHardenJDZhangXPIron(III) porphyrin catalyzed aziridination of alkenes with bromamine-T as nitrene sourceOrg Lett20046121907191015176780
- LiuYWuNDongJHfq is a global regulator that controls the pathogenicity of Staphylococcus aureusPloS One20105919
- LeejaeSHasapLVoravuthikunchaiSPInhibition of staphyloxanthin biosynthesis in Staphylococcus aureus by rhodomyrtone, a new antibiotic candidateJ Med Microbiol201362Pt 342142823242641