 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Induction of ampC β-lactamase expression can often compromise antibiotic treatment and is triggered by several β-lactams (such as cefoxitin and imipenem) and by the β-lactamase inhibitor clavulanic acid. The novel β-lactamase inhibitor avibactam (NXL104) is a potent inhibitor of both class A and class C enzymes. The potential of avibactam for induction of ampC expression in Enterobacter cloacae was investigated by ampC messenger ribonucleic acid quantitation. Cefoxitin and clavulanic acid were confirmed as ampC inducers, whereas avibactam was found to exert no effect on ampC expression. Thus, avibactam is unlikely to diminish the activity of any partner β-lactam antibiotic against AmpC-producing organisms.
Keywords:
Introduction
Bacterial resistance to β-lactams and β-lactamase inhibitors is an ever-increasing problem that compromises their clinical utility. Among Gram-negative bacteria, the production of β-lactamases is the most frequent factor contributing to β-lactam resistance. Of particular concern are enzymes able to target the expanded-spectrum β-lactams, including the AmpC enzymes (class C cephalosporinases), the so-called extended-spectrum-β-lactamases (ESBL; classes A and D), and the carbapenemases, which hydrolyze most β-lactams, including the carbapenems (classes A, B, and D).Citation1
In order to restore their antibacterial activity against Gram-negative pathogens, β-lactams have been paired with inhibitors of β-lactamases. Those currently used in the clinical setting (clavulanate, tazobactam, and sulbactam) have a spectrum of inhibition essentially covering class A enzymes. All three marketed inhibitors contain a β-lactam core and share a similar mechanism of inhibition. They react with serine enzymes to form a covalent acyl-enzyme intermediate; opening of the four-member β-lactam ring is followed by considerable molecular rearrangement before hydrolysis to regenerate the active enzyme.Citation2
Avibactam (NXL104) is a non-β-lactam β-lactamase inhibitor that displays a broad-spectrum inhibition profile, with potent inhibition of class A, class C, and some class D enzymes. The inhibitor is characterized by high carbamylation efficiency and slow decarbamylation, resulting in a long half-life of the inactive covalent adduct.Citation3 In addition, the decarbamylation step results in regeneration of intact avibactam, and not hydrolysis.Citation4 Avibactam has little intrinsic antibacterial activity, but efficiently protects β-lactams from hydrolysis in a variety of class A, class C, and some class D-producing strains, including ESBL, Klebsiella pneumoniae carbapenemase (KPC), and OXA-48 producers.Citation5–Citation7
Many strains of the Enterobacteriaceae family, as well as some nonfermenters, such as Pseudomonas aeruginosa and Acinetobacter baumannii, encode a chromosomal AmpC β-lactamase, although the regulation of enzyme production is different among the various enzyme-producing strains. In Escherichia coli, enzyme expression is usually constitutive and low-level, whereas in other species, such as Enterobacter spp., Citrobacter freundii, Serratia spp., Morganella morganii, Providencia spp., or P. aeruginosa, it can be transiently induced to higher levels by several β-lactam compounds, with carbapenems and cephamycins generally having the highest induction potential.Citation8 Derepression can also occur by mutations favoring constitutive production of very high levels of β-lactamase. The mechanism of induction is complex, via a system involving AmpD, AmpR, AmpG, and intermediates in peptidoglycan recycling.Citation9
Induction of ampC does not necessarily correlate with a risk of clinical failure, particularly when the rate of bactericidality is high. However, the potential for ampC induction has to be carefully examined when considering administration of a β-lactamase inhibitor, because it can antagonize the antibacterial activity of its partner β-lactam.Citation10–Citation12 Indeed, the antibacterial activity of a given β-lactam with limited stability to AmpC is preserved, provided that its potential for ampC induction is low. In contrast, its activity would be compromised if combined with a β-lactamase inhibitor that induces significant AmpC production.
The aim of this study was to investigate the ability of avibactam to induce ampC expression in E. cloacae strains in vitro. Because β-lactamase/avibactam complexes are known to have a long half-life, it was not possible to evaluate AmpC induction by measuring directly the β-lactamase activity produced by avibactam-treated cells. Thus, it was measured by quantitation of cellular ampC messenger ribonucleic acid (mRNA). Cefoxitin and clavulanate were used as reference ampC inducers.
Materials and methods
Bacterial strains and susceptibility testing
E. cloacae isolates used in this study were obtained from the Novexel culture collection, originally collected from a variety of clinical or laboratory sources. Minimal inhibitory concentration (MIC) determinations were performed according to the Clinical and Laboratory Standards Institute broth-microdilution methods using cation-adjusted Mueller–Hinton broth.Citation13 MIC values were measured for cefoxitin (Sigma-Aldrich, St Louis, MO, USA) and ceftazidime (Novartis, Basel, Switzerland); the latter was tested alone or in association with clavulanate (US Pharmacopeial Convention, Rockville, MD, USA) or avibactam, at a constant inhibitor concentration of 4 mg/L.
Induction experiments
Bacterial strains were grown overnight at 37°C in Luria–Bertani broth (Interchim, Montlugon, France), then diluted to an optical density (OD600 nm) value of 0.1 and incubated with shaking for 2–4 hours to reach midlog-growth phase. At this point, the test inducer (cefoxitin, clavulanate, or avibactam) was added at the appropriate concentration (8, 16, 32, or 64 μg/mL), whereas control cultures were grown in the absence of inducer. Approximately 5 × 108 cells were sampled for RNA extraction just before addition of inducer, and at timed intervals thereafter up to 6 hours. Each induction experiment was performed at least three times for all three E. cloacae strains.
Reverse-transcription polymerase chain reaction
Total cellular RNA was extracted with an RNeasy RNA Protect Mini Kit (Qiagen, Venlo, the Netherlands), and residual deoxyribonucleic acid (DNA) was eliminated by treatment with an RNAse-free DNAse Set (Qiagen). Analysis of RNA integrity and total RNA quantification was performed using the Agilent 2100 RNA bioanalyzer and the Nano 6000 kit (Agilent Technologies, Santa Clara, CA, USA).
Polymerase chain reaction (PCR) primers were designed with Primer Express software for ampC (forward 5′-TGGCGTATCGGGTCAATGT-3′; probe 5′-TCAGGGTCTGGGCTGGGAGATGC-3′; reverse 5′-CCTCCACGGGCCAGTTG-3′) and for rplS (forward 5′-CAGGTGACACCGTGGAAGTG-′; probe 5′-AAGTATGGGTTGTTGAAGGTTCCAA-3′; reverse 5′-CGAATGCCTGCAGACGTTT-3′). The probe primers were modified by addition of 6-FAM (6-carboxy-fluorescein) at the 5′ end and TAMRA (6-carboxy-tetramethyl-rhodamine) at the 3′ end.
Real-time PCR (RT-PCR) reactions were carried out in the ABI Prism® 7000 sequence-detection system (Life Technologies, Carlsbad, CA, USA) using a Quantitect Probe RT-PCR kit (Qiagen). Individual reactions were set up in triplicate for either ampC or rpsL genes, according to the manufacturer’s recommendations. Briefly, complementary DNA was syn-thesized from 0.5 ng of RNA using Moloney murine leuke-mia virus reverse transcriptase and 0.7 μM of each primer; reverse transcription was carried out at 50°C for 30 minutes. PCR conditions were as follows: initial activation of DNA polymerase at 95°C for 15 minutes, and PCR for 40 cycles at 95°C for 15 seconds, 60°C for 60 seconds. Absence of genomic DNA contamination was verified for each RNA preparation by running the assay in the absence of reverse transcriptase. Data were analyzed using Sequence Detection 2.0 software (Life Technologies). To correct for differences in the amount of starting material, the ribosomal E. cloacae rplS gene (encoding ribosomal protein 19) was chosen as a housekeeping reference gene. Values obtained were then normalized to that of ampC from E. cloacae strain P99 for measurement of basal expression, or to that of ampC in the test strain before induction. Relative quantitation was carried out by using the 2−ΔΔCT method, as recommended by the manufacturer.Citation14 Normalized ampC expression in culture 2 relative to that in culture 1 was calculated as follows:
β-Lactamase activity assays
Crude bacterial extracts were prepared by vortexing bacterial cells with glass beads in 100 mM phosphate buffer pH 7 containing 0.1 mg/mL of bovine serum albumin and 2% v/v glycerol (about 1011 bacterial cells/mL). β-Lactamase activity was measured using a spectrophotometer at 485 nm for 15 minutes using 180 µL of crude cell lysate at appropriate dilution, and 20 µL of 1 mM nitrocefin (Oxoid SR112C). Results were expressed as initial reaction rates (ΔA485 nm/minute) per 106 cells or per milligram of protein.
Results
Both cefoxitin and ceftazidime are good AmpC substrates; cefoxitin is also a good ampC inducer, whereas ceftazidime has limited potential for induction. The differential activity of cefoxitin and ceftazidime can therefore be used to infer the presence of an inducible ampC gene, as strains without significant levels of AmpC enzyme remain susceptible to cefoxitin.Citation9,Citation15 In order to select potentially inducible strains for this study, three E. cloacae strains (293LA2, 293HT107, and 293UC1) were chosen on the basis of resistance to cefoxitin and susceptibility to ceftazidime. The E. cloacae P99 strain was included in this study as a reference, having a cefoxitin-resistant (cefoxitin-R) and ceftazidime-R phenotype (stably derepressed high-level AmpC producer). The MIC values obtained for these four strains are shown in . As expected, owing to its spectrum of coverage limited to class A enzymes, clavulanate had no effect on the high ceftazidime MIC value for the P99 strain; in contrast, avibactam reduced ceftazidime MIC to a susceptible level in this strain.
Table 1 Susceptibility to antibiotics, basal expression of ampC mRNA and β-lactamase activity in Enterobacter cloacae strains
Induction of β-lactamases is most frequently assessed by assaying β-lactamase activity using spectrophotometric assays of nitrocefin hydrolysis, in the presence or absence of an inducer. However, this is not technically possible when testing the induction potential of a compound that can form highly stable complexes with AmpC enzymes (half-life around 7 days for avibactam/P99 AmpC complex).Citation5 Therefore, induction was measured by quantitation of ampC transcripts using RT-PCR. However, β-lactamase enzymatic activity was measured using nitrocefin in parallel with RT-PCR in the experiments that did not involve exposure to β-lactamase inhibitors. Basal expression of ampC mRNA in those three E. cloacae strains was compared to that of derepressed AmpC P99 and found to be 150- to 300-fold lower (). Concomitantly, basal β-lactamase activity in crude cell extracts reported using the nitrocefin substrate was also higher (1,000- to 1,800-fold) in P99 than in the three selected E. cloacae strains. Basal levels of ampC transcripts and β-lactamase activity were therefore fully consistent with the susceptibility/resistance phenotype of the strains.
The inducibility of β-lactamase activity was studied in the presence of 1–32 mg/L cefoxitin. The three E. cloacae strains tested showed similar induction profiles: ampC mRNA peaked at 1–2 hours following induction, then slowly declined to reach basal levels at 4–6 hours. shows the kinetics of one representative experiment with the strain 293HT107 treated with cefoxitin at 1 or 2 mg/L. Increased mRNA concentrations were detectable soon after incubation start (as soon as 10 minutes; data not shown) and peaked after 1–2 hours of incubation. β-Lactamase activity was delayed slightly when compared to ampC mRNA and continuously increased throughout the 4 hours of incubation with cefoxitin. When treated with 16–32 mg/L of cefoxi-tin, the maximal ampC transcriptional level after 2 hours of culture was between 100 and 200 times the basal level of both 293HT107 () and 293LA2 () strains, and around 50 times the basal level of the 293UC1 strain ().
Figure 1 Kinetics of ampC transcription and β-lactamase activity in Enterobacter cloacae 293HT107. E. cloacae strain 293HT107 was incubated with cefoxitin at 1 mg/L (triangles), 2 mg/L (squares), or in control medium (open circles). After 0.5, 1, 2, and 4 hours of culture, ampC messenger ribonucleic acids (mRNA) were quantified by real-time polymerase chain reaction (continuous lines; values show fold change in comparison with transcription level before incubation), and β-lactamase activity was measured spectrophotometrically (dashed lines).
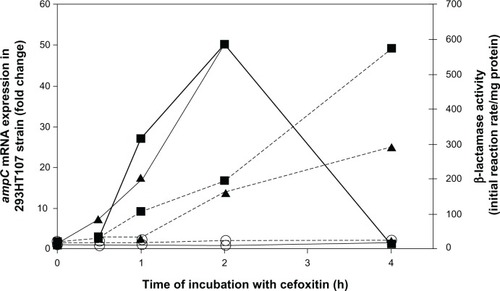
Figure 2 (A–I) Potential for ampC induction of cefoxitin, clavulanate, and avibactam. Enterobacter cloacae strains 293UC1 (A–C), 293LA2 (D–F), and 293HT107 (G–I) were incubated with cefoxitin (A, D and G), clavulanate (B, E and H), or avibactam (C, F and I). Inducers were used at various concentrations: 8 mg/L (squares), 16 mg/L (triangles), 32 mg/L (filled circles), or 64 mg/L (open circles); control cultures are shown with dashed lines. ampC messenger ribonucleic acids were quantified by real-time polymerase chain reaction after 2, 4, and 6 hours of culture.
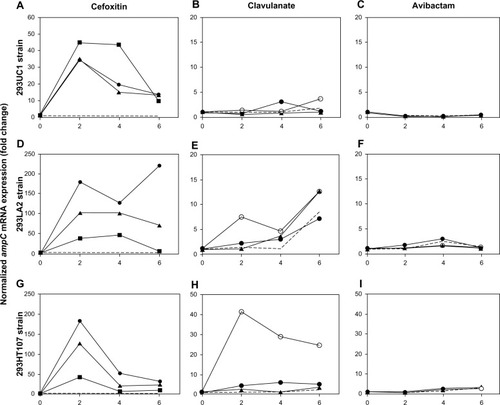
The potential of avibactam and clavulanate for induction of ampC expression was evaluated on the three E. cloacae strains at 16–64 mg/L. Clavulanate had no significant effects on 293UC1 and 293LA2 strains during the 6-hour incubation period (), whereas it was a moderate ampC inducer for 293HT107 with about a 40-fold increase of ampC mRNA at 64 mg/L after 2 hours of incubation (). In contrast, avibactam had no detectable effect on ampC mRNA levels in the three strains tested (). At the concentrations used for induction studies, avibactam had no effect on the growth of the bacterial strains tested, as testified by the OD values measured at each time point.
Discussion
Enterobacter spp. are recognized to be among the most common nosocomial pathogens, with current resistance rates presenting a serious therapeutic dilemma. Resistance through overexpression of AmpC can occur in the vast majority of strains possessing a chromosomally encoded cephalosporinase, and ampC induction is recognized as a widespread resistance mechanism. In a study examining 200 clinical isolates of P. aeruginosa, Enterobacter spp., Citrobacter spp., and Serratia spp., it was shown that 85% of the collected strains showed inducible AmpC production, of which 11% were stably derepressed and only 3% were not induced by either cefoxitin or imipenem.Citation16 Approximately 12% of hospital strains of the European Meropenem Yearly Susceptibility Test Information Collection (MYSTIC) program in the years 1997–2000 were due to potential AmpC-producing strains of Enterobacter spp., Citrobacter spp., and S. marcescens, in which 28% represented stably derepressed AmpC-producing phenotypes.Citation17
Much of what is known about AmpC regulation is from studies in E. coli, C. freundii, and E. cloacae; the induction mechanism in response to exposure to certain β-lactams is complex and closely linked to the peptidoglycan-recycling pathway.Citation9 Different effector proteins and regulation mechanisms have been recently evidenced for P. aeruginosa ampC induction, suggesting that the process is more complex in that species and distinct from the current paradigm established following studies of Enterobacteriaceae species.Citation18
β-lactams differ in their inducing abilities, with carbapen-ems and cephamycins having the highest potential.Citation8 The clavu-lanate β-lactamase inhibitor is also an ampC inducer, and was shown in vitro to antagonize the antibacterial activity of various β-lactams.Citation10,Citation11 In this context, the potential for induction of the new β-lactamase inhibitor avibactam was evaluated. At sub-MIC concentrations, cefoxitin induced a major dose-dependent synthesis of ampC in all three E. cloacae strains tested here, whereas clavulanate triggered synthesis of ampC mRNA in two out of the three strains, at the highest concentration tested (64 mg/L). In contrast, in the same range of concentrations, avibactam had no effect on cellular ampC mRNA concentration in any of the three E. cloacae strains during the 6-hour incubation period. From these initial findings, it is concluded that there is little likelihood of antagonism between β-lactam antibiotics and the novel β-lactamase inhibitor avibactam in Enterobacter spp. Questions remain about other bacterial species producing inducible chromosomal AmpC enzymes, and will be the focus of future studies.
Avibactam is the first compound of a diazabicyclo-octane series. In contrast with the inhibitors currently available (clavulanate, tazobactam, and sulbactam), which all have relatively limited activity against the class C enzymes, avibactam is a potent inhibitor of AmpC β-lactamases.Citation3,Citation5 It is the first non-β-lactam β-lactamase inhibitor to advance to clinical development, currently undergoing Phase II–III clinical trials in combination with ceftazidime and with cef-taroline (http://www.clinicaltrials.gov). Ceftaroline, like some third-generation cephalosporins, is a weak inducer of AmpC enzymes at sub-MIC concentrations, resulting in a propensity to select AmpC-derepressed or AmpC-hyperinducible mutants.Citation19 Pairing ceftaroline with avibactam should thus be an effective strategy to limit the risk of selection of mutants, and to restore ceftaroline activity against AmpC-hyperproducing strains, as well as to other β-lactamase producers.
Acknowledgments
The authors are very grateful to Dr Ken Coleman for critical review of the manuscript.
Disclosure
This work was presented in abstract form at the International Congress of Antimicrobial Agents and Chemotherapy in 2006. The authors report no conflicts of interest in this work.
References
- PooleKResistance to β-lactam antibioticsCell Mol Life Sci2004612200222315338052
- DrawzSMBonomoRThree decades of β-lactamase inhibitorsClin Microbiol Rev20102316020120065329
- StachyraTPéchereauMCBruneauJMMechanistic studies of the inactivation of TEM-1 and P99 by NXL104, a novel non-β-lactam β-lactamase inhibitorAntimicrob Agents Chemother2010545132513820921316
- EhmannDEJahićHRossPLAvibactam is a covalent, reversible, non-β-lactam β-lactamase inhibitorProc Natl Acad Sci U S A2012109116631166822753474
- BonnefoyADupuis-HamelinCSteierVIn vitro activity of AVE1330A, an innovative broad-spectrum non-β-lactam β-lactamase inhibitorJ Antimicrob Chemother20045441041715254025
- LivermoreDMMushtaqSWarnerMMiossecCWoodfordNNXL104 combinations versus Enterobacteriaceae with CTX-M extended-spectrum β-lactamases and carbapenemasesJ Antimicrob Chemother2008621053105618689875
- StachyraTLevasseurPPéchereauMCIn vitro activity of the β-lactamase inhibitor NXL104 against KPC-2 carbapenemase and Enterobacteriaceae expressing KPC carbapenemasesJ Antimicrob Chemother20096432632919493866
- JonesRNImportant and emerging β-lactamase-mediated resistances in hospital-based pathogens: the AmpC enzymesDiagn Microbiol Infect Dis1998314614669635237
- JacobyGAAmpC β-lactamasesClin Microbiol Rev20092216118219136439
- KitzisMDFerréBCoutrotAIn vitro activity of combinations of β-lactam antibiotics with β-lactamase inhibitors against cephalosporinase-producing bacteriaEur J Clin Microbiol Infect Dis198987837882556277
- ListerPDGardnerVMSandersCCClavulanate induces expression of the Pseudomonas aeruginosa AmpC cephalosporinase at physiologically relevant concentrations and antagonizes the antibacterial activity of ticarcillinAntimicrob Agents Chemother19994388288910103195
- SandersCCSandersWEJrGoeringRVIn vitro antagonism of β-lactam antibiotics by cefoxitinAntimicrob Agents Chemother1982219689756981376
- Clinical and Laboratory Standards InstitutePerformance Standards for Antimicrobial Susceptibility Testing: Eighteenth International SupplementWayne (PA)CLSI2010
- LivakKJSchmittgenTDAnalysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT methodMethods20012540240811846609
- LivermoreDMWinstanleyTGShannonKPInterpretative reading: recognizing the usual and inferring resistance mechanisms from resistance phenotypesJ Antimicrob Chemother200148Suppl 18710211420342
- DunneWMJrHardinDJUse of several inducer and substrate antibiotic combinations in a disk approximation assay format to screen for AmpC induction in patients isolates from Pseudomonas aeruginosa, Enterobacter spp., Citrobacter spp., and Serratia sppJ Clin Microbiol2005435945594916333080
- PfallerMAJonesRNAntimicrobial susceptibility of inducible AmpC β-lactamase-producing Enterobacteriaceae from the Meropenem Yearly Susceptibility Test Information Collection (MYSTIC) Programme, Europe 1997–2000Int J Antimicrob Agents20021938338812007846
- KongKFAguilaASchneperLMatheeKPseudomonas aeruginosa β-lactamase induction requires two permeases, AmpG and AmpPBMC Microbiol20101032821192796
- MushtaqSLivermoreDMAmpC induction by ceftarolineJ Antimicrob Chemother20106526627019996139