Abstract
Background
Autophagy is an evolutionary conserved important process for the turnover of intracellular substances in eukaryotes and is closely related to the development of atrial fibrillation (AF). The aim of this study is to identify and validate potential autophagy-related genes (ARGs) of AF through bioinformatics analysis and experimental validation.
Methods
We downloaded two data sets from the Gene Expression Omnibus (GEO) database, GSE14975 and GSE31821. After merging the data of the two microarrays, adjusting the batch effect, and integrating the differentially expressed genes (DEGs) with ARGs to obtain differentially expressed autophagy-related genes (DEARGs). Functional and pathway enrichment analyses were carried out based on Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). Use the STRING database to construct a protein–protein interaction (PPI) network. Finally, mRNA expression levels of DEARGs were validated in right atrial tissue samples from AF patients and non-AF controls by qRT-PCR.
Results
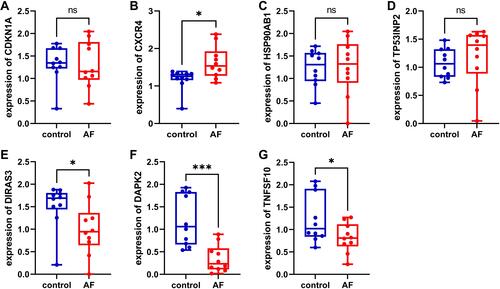
Through bioinformatics analysis, we finally identified 11 DEARGs (CDKN1A, CXCR4, DIRAS3, HSP90AB1, ITGA3, PRKCD, TP53INP2, DAPK2, IFNG, PTK6, and TNFSF10) in AF using [log2 (fold change)] > 0.5 and P < 0.05. In the pathway enrichment analysis, the most significantly enriched pathway was the autophagy pathway. The results of validation showed that the expression levels of CXCR4, DAPK2, and TNFSF10 corroborating with our computational findings, and the results were statistically significant (P<0.05).
Conclusion
Our study demonstrates that these 11 potential crucial ARGs, especially CXCR4, DAPK2, and TNFSF10, may be potential biomarkers and therapeutic targets in AF, which will help the personalized treatment of AF patients.
Introduction
Atrial fibrillation (AF) is the most common clinically sustained cardiac rhythm disorder. There is a high prevalence of AF all over the world, the highest in North America, Europe, China, and Southeast Asia, with approximately 270–360 cases per 100,000 people.Citation1 Due to the lack of understanding of the pathogenesis of AF, the current therapies available can only control AF in a short period of time, but cannot cure the disease, which brings a serious burden to the lives of AF patients. The pathogenesis of AF includes atrial remodeling, electrophysiological mechanisms, and the role of the autonomic nervous system, etc. Accumulating evidence has shown that the pathogenesis of AF involves a variety of biological processes, including apoptosis, immunoregulation, and autophagy.Citation2–Citation4 Among these biological processes, autophagy plays an important role in the development of AF.
Autophagy is the main intracellular degradation system. In the process of autophagy, the cytoplasm is encapsulated in a double-membrane structure of autophagosomes, and autophagosomes fuse with lysosomes to form autophagolysosomes, which are degraded in autophagosomes.Citation5 A lot of evidence shows that autophagy response is closely related to the occurrence and development of cardiovascular disease.Citation6–Citation9 For example, ischemia injury activates autophagy of cardiomyocytes, enabling cardiomyocytes to cope with nutritional stress and improving cell survival rate during ischemia-reperfusion injury.Citation10 For coronary heart disease, enhanced autophagy can not only protect the myocardium against ischemia but also prevent the remodeling of the heart after ischemia.Citation11 In recent years, there are also some studies on autophagy and AF.Citation12–Citation14 Observation of 170 patients in sinus rhythm who had undergone elective coronary artery bypass grafting, Garcia et al found that impaired autophagy plays an important role in the occurrence of postoperative AF.Citation15 Yuan et al indicated that there is AMPK-dependent autophagy in the occurrence of AF,Citation4 their subsequent research found that autophagy can induce atrial electrical remodeling through ubiquitin-dependent selective degradation of Cav1.2.Citation16 However, the understanding of the role of autophagy in the occurrence and development of AF is far from enough.
The purpose of this study is to deeply understand the potential clinical application value of autophagy-related genes (ARGs) in AF through the Gene Expression Omnibus (GEO) database using bioinformatic methods. Although Liu et al have previously conducted bioinformatics analysis of ARGs and AF, they only analyzed one dataset and did not verify the results.Citation17 In this study, we analyzed two datasets and verified the finally identified differential ARGs in patients with AF and non-AF individuals, which further improved the reliability of the results.
Materials and Methods
Data Collection and Preprocessing
GSE14975 and GSE31821 (10 samples in GSE14975 and 6 samples in GSE31821) were downloaded from GEO (http://www.ncbi.nlm.nih.gov/geo) and the sample platforms used were GPL570 (HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array.Citation18 The two data sets were selected because they were both derived from the patient’s atrial tissue and the same sequencing platform.
The data were preprocessed as follows: to annotate the data, the probe names were converted into gene names with ActivePerl 5.28.1 software (https://www.activestate.com/products/perl/downloads/). Then we generated a synthetic dataset from GSE14975 and GSE31821 datasets. For the new dataset, eliminate probes without related gene symbols, and calculate the average expression value of those gene symbols with multiple probes. The “sva” package of R software was used to remove batch-effects for new dataset batch correction.
Screening of Differentially Expressed Genes (DEGs)
The DEGs were screened with R software limma package version 3.44.3 (http://bioconductor.org/packages/release/bioc/html/limma.html) under the criteria of [log2 (fold change)] >0.5 and P <0.05.
Protein-Protein Interaction (PPI) Network and Functional Analysis of DEGs
PPI networks of DEGs were analyzed using the STRING online tool (STRING database, version 11.0; https://string-db.org/cgi/input.pl?sessionId=1fJdSiXTVpOdandinput_page_show_search=on) to further predict protein functional associations and protein-protein interactions.Citation19 It might provide insights into the underlying molecular mechanism of the initiation and progression of diseases. An interaction with a confidence score >0.70 was considered statistically significant. The specific process of DEGs function and pathway enrichment analysis is consistent with that described by differentially expressed ARGs (DEARGs).
Autophagy-Related Genes
We obtained a total of 232 ARGs from the Human Autophagy-dedicated Database (HADb, http://autophagy.lu/clustering/index.html), which provides a more detailed list of human genes involved in autophagy.
Identification of DEARGs
Data were further analyzed by R software, taking the intersection of DEGs expression profile and 232 ARGs to identify DEARGs.
Functional and Pathway Enrichment Analysis of DEARGs
Through Gene Ontology (GO) enrichment analysis, we can comprehensively understand the biological process, cellular component, and molecular function of DEARGs enriched. In this study, the bohao online enrichment tool (http://enrich.shbio.com/index/ga.asp) was utilized to perform GO enrichment analysis on DEARGs. Terms of which P <0.05 were statistically significant.
The identified DEARGs were also analyzed by pathway enrichment in the Kyoto Encyclopedia of Genes and Genomes (KEGG) to find relevant important pathways. ClusterProfiler version 3.16.1 (https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html) was used to perform KEGG pathway enrichment analysis in R software.Citation20,Citation21 Pathways of which P <0.05 were statistically significant.
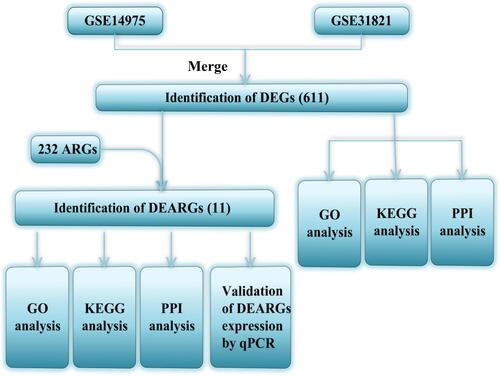
A detailed flow chart for the specific process of analysis was shown in .
Figure 1 The flow chart shows the design of the present study.
AF Patients and Non-AF Controls
A total of 10 AF patients and 10 non-AF controls were obtained from the Fuwai Yunnan Cardiovascular Hospital between April 2021 and July 2021. This study included patients who underwent thoracotomy due to mitral valve replacement and/or atrial septal defect. Patients with AF (defined as persistent atrial fibrillation lasting more than 7 days) were assigned to AF, while those without AF were assigned to the control group. Patients were excluded if they had thyroid dysfunction and active rheumatism. This study was approved by the Ethics Committee of Fuwai Yunnan Cardiovascular Hospital (Ethical Application Ref: IRB2021-BG-006). Written informed consent was obtained from all individual participants included in the study. Right atrial tissue was collected from patients who participated in the study.
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from right atrial tissue with TRIzol Reagent (Life Technologies, CA, USA). Reverse transcription was conducted using SureScript-First-strand-cDNA-synthesis-kit (GeneCopoeia, Guangzhou, China). qPCR was conducted using a BlazeTaq™ SYBR® Green qPCR Mix 2.0 kit (GeneCopoeia, Guangzhou, China) following the instructions. The thermocycling conditions were as follows: initial activation at 95°C for 30 s, followed by 40 cycles at 95°C for 10 s, 60°C for 20 s and 72°C for 30 s. GAPDH was used as the internal reference for the mRNA for data normalization. The relative expression was calculated by 2−ΔΔCt method. Primers are available in .
Table 1 Primer Sequences for qRT- PCR
Statistical Analysis
The statistical analyses were performed using GraphPad Prism 9.0 software (San Diego, CA, USA). Real-time gene expression levels of our tissue samples were compared using Student’s t-test. P < 0.05 was considered to indicate statistical significance.
Results
Data Preprocessing and Differential Expression Analysis
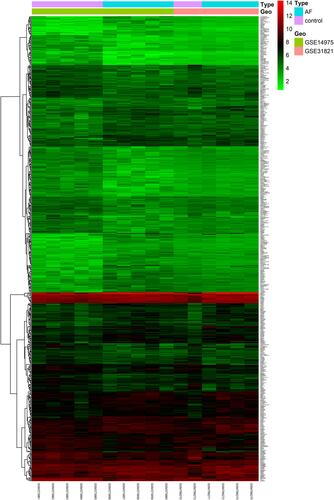
A total of 54,675 probes were obtained from GSE14975 and GSE31821. After preprocessing, 21,644 genes were identified. Considering the criteria for [log2 (fold change)] >0.5 and P <0.05, we finally obtained a total of 611 significant DEGs, of which 309 were up-regulated and 302 were down-regulated. The clustering heatmap is shown in .
PPI Network and Functional GO Terms and Pathway Enrichment Analyses of DEGs
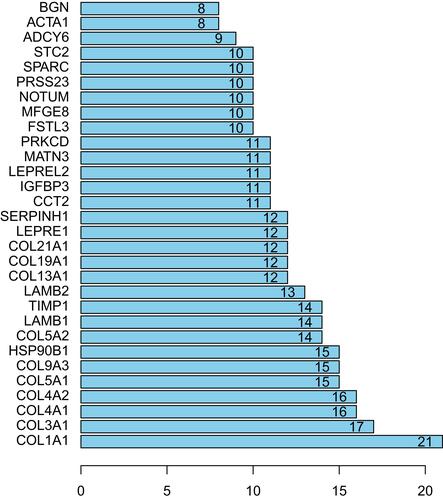
Altogether, 202 nodes and 389 interaction pairs were identified in the PPI network (). According to the view that highly connected genes were noted to possibly play important roles in diseases, we calculated the connectivity between the nodes through R software and the results are displayed in . Here, the first 6 nodes are all members of the collagen family, including collagen type I alpha 1 chain (COL1A1, degree =21), collagen type III alpha 1 chain (COL3A1, degree =17), collagen type IV alpha 1 chain (COL4A1, degree =16), collagen type IV alpha 2 chain (COL4A2, degree =16), collagen type V alpha 1 chain (COL5A1, degree =15) and collagen type IX alpha 3 chain (COL9A3, degree =15) are considering as hub genes in related to AF maintaining.
Figure 3 PPI network of DEGs between control samples and atrial fibrillation samples.
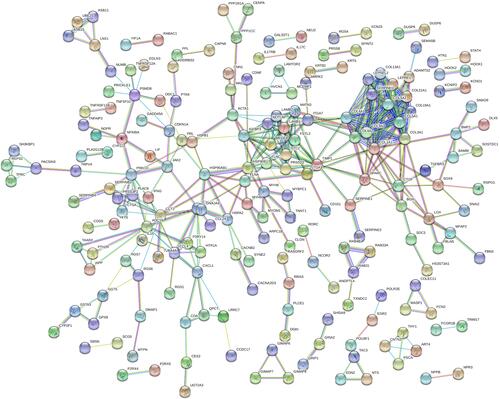
Figure 4 Top 30 nodes of PPI networks of DEGs between control samples and AF samples.
To investigate the biological effects of DEGs, we performed GO and KEGG functional enrichment analyses, the top 3 GO terms related biological processes were collagen catabolic process (enrich factor: 6.62; P-value: 3.148e-07), collagen fibril organization (enrich factor: 7.92; P-value: 1.713e-05) and negative regulation of cell-cell adhesion (enrich factor: 7.00; P-value: 3.780e-07), the results are shown in the .
Figure 5 GO (A) analysis shows the biological processes, cellular components, and molecular functions involved in DEGs. Bar plot (B) and dot plot (C) show KEGG pathway enrichment of DEGs.
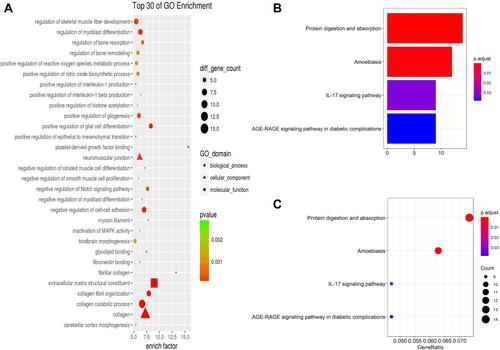
KEGG pathway analysis data appear in and . The results suggest that DEGs were significantly enriched in pathways of protein digestion and absorption (P-value: 4.24e-08), amoebiasis (P-value: 4.72e-06), and IL-17 signaling pathway (P-value: 0.00037).
Identification of DEARGs
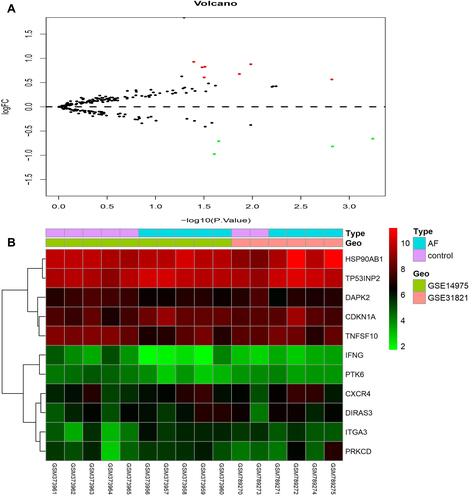
Using the DEGs identified in the previous step and extracted the expression values of 232 ARGs. A total of 11 DEARGs were identified (see and for details), including 7 up-regulated genes (CDKN1A, CXCR4, DIRAS3, HSP90AB1, ITGA3, PRKCD, TP53INP2) and 4 down-regulated genes (DAPK2, IFNG, PTK6, TNFSF10). Besides, a heat map was visualized to show the relative expression pattern of 11 DEARGs between control samples and AF samples ().
Table 2 The DEARGs of Merged Data of 611 DEGs and 232 ARGs with the Use of Criteria of Log2 (Fold Change) >0.5 and P <0.05
Figure 6 Volcano map (A) shows DEARGs, with red dots representing up-regulated genes, green dots representing down-regulated genes, and the remaining black dots representing no differences gene. Heat map (B) of 11 DEARGs in AF samples and control samples.
PPI Network and Functional Enrichment of the DEARGs
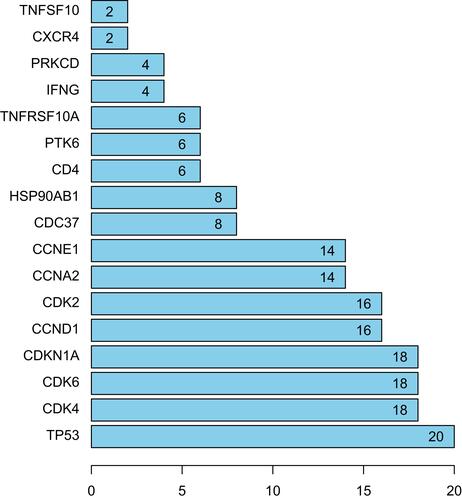
The PPI network based on DEARGs consisted of 17 nodes and 90 interaction pairs ( and ). The top 8 nodes are mostly proteins related to cell cycle regulation, respectively TP53, CDK4, CDK6, CDKN1A, CCND1, CDK2, CCNA2, and CCNE1.
Figure 7 PPI network of DEARGs between control samples and atrial fibrillation samples.
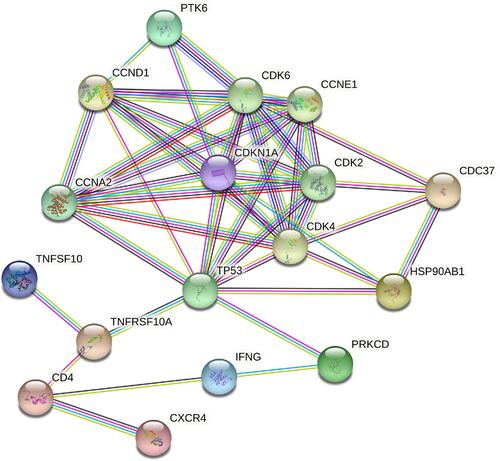
Figure 8 Top 17 nodes of PPI networks of DEARGs between control samples and AF samples.
To further explore the biological functions of the DEARGs, functional enrichment and pathway analyses were performed and the results are presented in –.
Figure 9 GO (A) analysis shows the biological processes, cellular components, and molecular functions involved in DEARGs. Bar plot (B) and dot plot (C) show KEGG pathway enrichment of DEARGs.
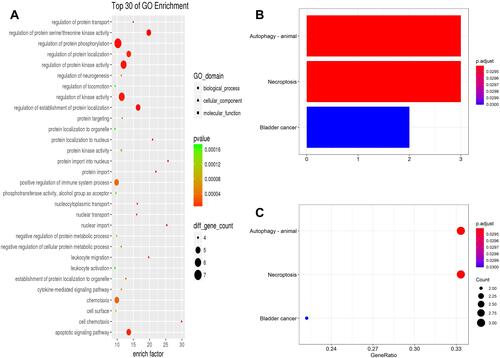
GO enrichment showed that changes in the biological process (BP) of DEARGs were mainly enriched in the protein phosphorylation, protein kinase activation, and apoptotic signaling pathway. (, ).
Table 3 Significant Enriched GO Terms and Pathways of DEARGs
The results of KEGG enrichment analysis showed that pathways of DEARGs mainly involve pathways in autophagy, necroptosis, and bladder cancer ( and ). It is worth mentioning that it is directly enriched into autophagy, which proves the correctness of the analysis results.
Validation of DEARGs Expression in AF Patients and Non-AF Controls
To confirm the reliability of the bioinformatics analysis, we validated DEARGs in patients with AF and control by qRT- PCR. The clinicopathological variables of cases and controls are summarized in . Similar to the results of mRNA microarray in AF tissue samples, the expression level of CXCR4 was found to be significantly up-regulated (), the expression levels of DAPK2 and TNFSF10 were found to be down-regulated ( and ). However, the expression levels of CDKN1A, HSP90AB1, and TP53INP2 showed no significant difference between AF patients and non-AF controls (–). Furthermore, we found that DIRAS3 was significantly down-regulated in AF patients (), which did not correlate with our bioinformatics analysis. IFNG, ITGA3, PRKCD, and PTK6 were not detected by qRT- PCR due to their low expression levels.
Table 4 Summary of Clinicopathological Variables of Cases and Controls
Discussion
The pathogenesis of AF includes atrial remodeling, electrophysiological mechanisms, and the role of the autonomic nervous system, etc. Among all of them, atrial remodeling is most closely associated with AF. It is well-known that myocardial fibrosis is the most important part of atrial remodeling. When the content of fiber components (collagen fiber) in myocardial tissue is higher, AF is more likely to persist. In this study, PPI network and GO analysis of DEGs also showed that collagen family and collagen catabolic process play a key role in the pathogenesis of AF, especially type I and III collagen, have been confirmed that their metabolic changes are significantly related to AF.Citation22 With the in-depth research in recent years, increasing evidence has demonstrated that autophagy may be involved in the regulation of myocardial fibrosis.Citation23–Citation25 Chikusetsu saponin IVa can activate autophagy through AMPK/mTOR/ULK1 pathway to reduce isoproterenol-induced myocardial fibrosis.Citation23 Calhex231 may ameliorate myocardial fibrosis by inhibiting autophagy-mediated activation of NLRP3 inflammasome.Citation26 However, extensive validation is needed to improve our understanding of autophagy in the pathogenesis of AF.
Autophagy is strongly linked to the development of AF, but the mechanisms involved are complex. The study of Hu et al has directly pointed out that, through in vivo and in vitro studies, they found that quercetin can inhibit the expression of miR-223-3p, while enhancing the expression of FOXO3 and activating the autophagy pathway, which significantly inhibits Myocardial fibrosis in AF.Citation27 But, there have been no studies using bioinformatics to explore the role of ARGs in AF and to conduct experimental verification. In the present study, we used bioinformatics tools to analyze the integrated data of gene expression profiles from two GEO datasets to identify key ARGs related to the therapeutic targets of AF patients. We found that 11 ARGs (CDKN1A, CXCR4, DIRAS3, HSP90AB1, ITGA3, PRKCD, TP53INP2, DAPK2, IFNG, PTK6, TNFSF10) under the criteria of [log2 (fold change)] >0.5 and P <0.05 were differentially expressed in AF patient myocardial tissue samples. Some of these ARGs of AF have been previously studied. For example, the expression of CXCR4 is upregulated in patients with chronic AF, leading to atrial remodeling.Citation28 This result is consistent with the results of our bioinformatics analysis and experimental verification. We intend to explore more potential ARGs of AF in the future.
The STRING database was used to identify the eight nodes with the greatest degree of network connection (TP53, CDK4, CDK6, CDKN1A, CCND1, CDK2, CCNA2, and CCNE1). These targets are mainly regulatory factors related to inflammation, DNA replication and repair, cellular senescence, cell cycle, and so forth.Citation29–Citation32 Myocardial fibrosis is one of the important mechanisms for the occurrence of AF, and these proteins may indirectly participate in the regulation of AF through myocardial fibrosis. The greatest number of node networks (n=20) was TP53. Tumor protein p53 (TP53) is a critical protein involved in the process of cell cycle, apoptosis, senescence, and DNA repair. Overactivation of TP53 pathway in cardiomyocytes induces myocardial fibrosis.Citation33 Similar to the studies conducted by Chen et al, the TP53 signaling pathway is activated in ventricular arrhythmias in dilated cardiomyopathy.Citation34 Therefore, TP53 may be an important target gene of AF. CDK4, CDK6, CDKN1A, CDK2, and CCNA2 are important proteins involved in the regulation of the mammalian cell cycle. The present study shows that CDK4/6 inhibitors could delay the progression of bleomycin-induced pulmonary fibrosis.Citation35 MicroRNA-1 can inhibit the proliferation of cardiac fibroblasts by directly acting on CDK6.Citation36 Moreover, activation of AMPK further inhibited CDK2 and cyclin E complexes by up-regulating CDKN1A expression, ultimately suppressing the progression of cardiac fibrosis.Citation37 Overexpression of CCNA2 also alleviates myocardial fibrosis in a porcine model of myocardial infarction.Citation38 These studies provide a convincing framework for the development of therapies to alleviate myocardial fibrosis based on cardiomyocyte cycle regulation. However, there exists no study to investigate the relationships between CCND1, CCNE1 and myocardial fibrosis.
The potential biological functions of these DEARGs were also analyzed by GO and KEGG enrichment analysis. Functional enrichment analysis in the present study indicated that the GO terms (biological process) were mainly enriched in protein phosphorylation regulation, protein kinase activity, and kinase activity. KEGG pathway enrichment analysis showed that PRKCD, TP53INP2, and DAPK2 were enriched in the autophagy pathway. Some published articles confirmed that autophagy can affect the progress of AF. One paper mentioned that autophagy can promote the occurrence and persistence of AF through the degradation of L-type calcium channels.Citation16 One other study found osteopontin induces the activation of AKT/mTOR, inhibits autophagy, aggravates the AF.Citation13 These findings suggest that the study of ARGs and AF may reveal the pathogenesis of AF.
Based on the bioinformatics analysis results, we further identified the expression level of DEARGs in our clinical samples by qRT-PCR. According to the results of qRT- PCR, the expression levels of CXCR4, DAPK2, and TNFSF10 were in accordance with the bioinformatics analysis results from mRNA microarray. Among them, CXCR4 has been confirmed to be closely associated with the occurrence and maintenance of AF. It was found that CXCR4 expression is up-regulated in patients with chronic AF with mitral valve disease.Citation28 Inhibition of CXCL12/CXCR4 axis can reduce the recruitment of inflammatory cells and suppress the hyperactivation of atrial ERK1/2 and AKT/mTOR signaling pathways, thereby alleviating AF.Citation39 As previously mentioned, myocardial fibrosis is one of the important pathogenesis of AF, and CXCR4 also plays a key regulatory role in myocardial fibrosis. It was found that CXCR4 antagonist significantly reduced the expression of collagen I mRNA and alleviated myocardial fibrosis in experimental dilated cardiomyopathy mice.Citation40 In addition, study on DAPK2, TNFSF10 and AF is not reported. It should be noted that due to the limited sample volume, we did not conduct experimental verification of differential protein abundance by analyzing gene expression. However, the differential protein identified in this study has also been found in other studies, as demonstrated by Liu et al, the protein expression level of CXCR4 in AF patients was significantly higher than that in patients with sinus rhythm.Citation39 Nevertheless, the current research on ARGs and AF is far from enough and needs further exploration.
In the current study, we discussed 11 potential crucial DEARGs involved in the occurrence and development of AF, suggesting that these genes may serve as potential biomarkers and therapeutic targets for AF. However, there are still some limitations in this study. Firstly, due to the small sample size of this study and the heterogeneity of AF, the interpretation of the study results needs to be cautious. In addition, the specific pathophysiological mechanisms of ARGs regulating the initiation and progression of AF need to be further studied. Finally, the working mechanism of these genes is not yet fully understood, so more evidence is needed to discover its biological basis.
Conclusion
In summary, we conducted the gene differential expression analysis, functional enrichment analysis, and protein-protein-interaction analysis of autophagy genes in AF, these results show that these DEARGs have great potential as biomarkers and therapeutic targets in AF. Future studies need to further validate the protein interactions identified by RT-PCR. In addition, the crucial genes CXCR4, DAPK2, and TNFSF10 may provide new possibilities for further identifying the susceptibility of AF and finding useful therapeutic targets.
Abbreviations
AF, atrial fibrillation; ARGs, autophagy-related genes; GEO, Gene Expression Omnibus; DEGs, differentially expressed genes; PPI, protein-protein interaction; DEARGs, differentially expressed ARGs; HADb, Human Autophagy-dedicated Database; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; qRT-PCR, quantitative real-time PCR; BP, biological process; BMI, body mass index; log2 FC, log2 (fold change); AveExpr, average expression; adj. P. Val, adjust P-value.
Data Sharing Statement
The data analyzed in the present study can be accessed on the GEO (https://www.ncbi.nlm.nih.gov/geo/) website.
Ethics Approval and Informed Consent
The study protocol was approved by the Ethics Committee of Fuwai Yunnan Cardiovascular Hospital (Ethical Application Ref: IRB2021-BG-006), Kunming, Yunnan, China. Informed consent forms were signed voluntarily by all individual participants included in the study.
Acknowledgments
We all authors sincerely acknowledge the contribution from the GEO depository.
Disclosure
The authors report no conflicts of interest in this work.
Additional information
Funding
References
- Joseph PG, Healey JS, Raina P, et al. Global variations in the prevalence, treatment, and impact of atrial fibrillation in a multi-national cohort of 153 152 middle-aged individuals. Cardiovasc Res. 2021;117(6):1523–1531. doi:10.1093/cvr/cvaa241
- Yu RB, Li K, Wang G, Gao GM, Du JX. MiR-23 enhances cardiac fibroblast proliferation and suppresses fibroblast apoptosis via targeting TGF-β1 in atrial fibrillation. Eur Rev Med Pharmacol Sci. 2019;23(10):4419–4424.
- Hu YF, Chen YJ, Lin YJ, Chen SA. Inflammation and the pathogenesis of atrial fibrillation. Nat Rev Cardiol. 2015;12(4):230–243. doi:10.1038/nrcardio.2015.2
- Yuan Y, Zhao J, Yan S, et al. Autophagy: a potential novel mechanistic contributor to atrial fibrillation. Int J Cardiol. 2014;172(2):492–494. doi:10.1016/j.ijcard.2014.01.027
- Liu F, Fang F, Yuan H, et al. Suppression of autophagy by FIP200 deletion leads to osteopenia in mice through the inhibition of osteoblast terminal differentiation. J Bone Miner Res. 2013;28(11):2414–2430. doi:10.1002/jbmr.1971
- Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat Rev Drug Discov. 2017;16(7):487–511.
- Bravo-San Pedro JM, Kroemer G, Galluzzi L. Autophagy and Mitophagy in Cardiovascular Disease. Circ Res. 2017;120(11):1812–1824. doi:10.1161/CIRCRESAHA.117.311082
- Ren J, Zhang Y. Targeting autophagy in aging and aging-related cardiovascular diseases. Trends Pharmacol Sci. 2018;39(12):1064–1076. doi:10.1016/j.tips.2018.10.005
- Ren J, Sowers JR, Zhang Y. Metabolic stress, autophagy, and cardiovascular aging: from pathophysiology to therapeutics. Trends Endocrinol Metab. 2018;29(10):699–711. doi:10.1016/j.tem.2018.08.001
- Mialet-Perez J, Vindis C. Autophagy in health and disease: focus on the cardiovascular system. Essays Biochem. 2017;61(6):721–732. doi:10.1042/EBC20170022
- Dong Y, Chen H, Gao J, Liu Y, Li J, Wang J. Molecular machinery and interplay of apoptosis and autophagy in coronary heart disease. J Mol Cell Cardiol. 2019;136:27–41. doi:10.1016/j.yjmcc.2019.09.001
- Wiersma M, Meijering RAM, Qi XY, et al. Endoplasmic reticulum stress is associated with autophagy and cardiomyocyte remodeling in experimental and human atrial fibrillation. J Am Heart Assoc. 2017;6(10):e006458. doi:10.1161/JAHA.117.006458
- Lin R, Wu S, Zhu D, Qin M, Liu X. Osteopontin induces atrial fibrosis by activating Akt/GSK-3β/β-catenin pathway and suppressing autophagy. Life Sci. 2020;245:117328. doi:10.1016/j.lfs.2020.117328
- Shingu Y, Takada S, Yokota T, et al. Correlation between increased atrial expression of genes related to fatty acid metabolism and autophagy in patients with chronic atrial fibrillation. PLoS One. 2020;15(4):e0224713. doi:10.1371/journal.pone.0224713
- Garcia L, Verdejo HE, Kuzmicic J, et al. Impaired cardiac autophagy in patients developing postoperative atrial fibrillation. J Thorac Cardiovasc Surg. 2012;143(2):451–459. doi:10.1016/j.jtcvs.2011.07.056
- Yuan Y, Zhao J, Gong Y, et al. Autophagy exacerbates electrical remodeling in atrial fibrillation by ubiquitin-dependent degradation of L-type calcium channel. Cell Death Dis. 2018;9(9):873. doi:10.1038/s41419-018-0860-y
- Liu A, Jia K, Liang H, Jin Q. Comprehensive analysis of autophagy-related genes and patterns of immune cell infiltration in valvular atrial fibrillation. BMC Cardiovasc Disord. 2021;21(1):132. doi:10.1186/s12872-021-01939-1
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi:10.1093/nar/gks1193
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi:10.1093/nar/gky1131
- Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi:10.1093/nar/28.1.27
- Xu J, Cui G, Esmailian F, et al. Atrial extracellular matrix remodeling and the maintenance of atrial fibrillation. Circulation. 2004;109(3):363–368. doi:10.1161/01.CIR.0000109495.02213.52
- Wang L, Yuan D, Zheng J, et al. Chikusetsu saponin IVa attenuates isoprenaline-induced myocardial fibrosis in mice through activation autophagy mediated by AMPK/mTOR/ULK1 signaling. Phytomedicine. 2019;58:152764. doi:10.1016/j.phymed.2018.11.024
- Lu C, Yang Y, Zhu Y, Lv S, Zhang J. An Intervention Target for Myocardial Fibrosis: autophagy. Biomed Res Int. 2018;2018:6215916. doi:10.1155/2018/6215916
- Lv S, Yuan P, Dong J, et al. QiShenYiQi pill improves the reparative myocardial fibrosis by regulating autophagy. J Cell Mol Med. 2020;24(19):11283–11293. doi:10.1111/jcmm.15695
- Liu W, Sun J, Guo Y, et al. Calhex231 ameliorates myocardial fibrosis post myocardial infarction in rats through the autophagy-NLRP3 inflammasome pathway in macrophages. J Cell Mol Med. 2020;24(22):13440–13453. doi:10.1111/jcmm.15969
- Hu J, Wang X, Cui X, Kuang W, Li D, Wang J. Quercetin prevents isoprenaline-induced myocardial fibrosis by promoting autophagy via regulating miR-223-3p/FOXO3. Cell Cycle. 2021;20(13):1253–1269. doi:10.1080/15384101.2021.1932029
- Wang XX, Zhang FR, Zhu JH, Xie XD, Chen JZ. Up-regulation of CXC chemokine receptor 4 expression in chronic atrial fibrillation patients with mitral valve disease may be attenuated by renin-angiotensin system blockers. J Int Med Res. 2009;37(4):1145–1151. doi:10.1177/147323000903700419
- Ham SW, Jeon HY, Jin X, et al. TP53 gain-of-function mutation promotes inflammation in glioblastoma. Cell Death Differ. 2019;26(3):409–425. doi:10.1038/s41418-018-0126-3
- Dai M, Boudreault J, Wang N, et al. Differential Regulation of Cancer Progression by CDK4/6 Plays a Central Role in DNA Replication and Repair Pathways. Cancer Res. 2021;81(5):1332–1346. doi:10.1158/0008-5472.CAN-20-2121
- Xu X, Shen X, Feng W, et al. D-galactose induces senescence of glioblastoma cells through YAP-CDK6 pathway [published online ahead of print, 2020 Sep 29]. Aging. 2020;12(18):18501–18521. doi:10.18632/aging.103819
- Liu K, Graves JD, Lee YJ, Lin FT, Lin WC. Cell Cycle-Dependent Switch of TopBP1 Functions by Cdk2 and Akt. Mol Cell Biol. 2020;40(8):e00599–19. doi:10.1128/MCB.00599-19
- Chen SN, Lombardi R, Karmouch J, et al. DNA Damage Response/TP53 Pathway Is Activated and Contributes to the Pathogenesis of Dilated Cardiomyopathy Associated With LMNA (Lamin A/C) Mutations. Circ Res. 2019;124(6):856–873. doi:10.1161/CIRCRESAHA.118.314238
- Haywood ME, Cocciolo A, Porter KF, et al. Transcriptome signature of ventricular arrhythmia in dilated cardiomyopathy reveals increased fibrosis and activated TP53. J Mol Cell Cardiol. 2020;139:124–134. doi:10.1016/j.yjmcc.2019.12.010
- Birnhuber A, Egemnazarov B, Biasin V, et al. CDK4/6 inhibition enhances pulmonary inflammatory infiltration in bleomycin-induced lung fibrosis. Respir Res. 2020;21(1):167. doi:10.1186/s12931-020-01433-w
- Valkov N, King ME, Moeller J, Liu H, Li X, Zhang P. MicroRNA-1-Mediated Inhibition of Cardiac Fibroblast Proliferation Through Targeting Cyclin D2 and CDK6. Front Cardiovasc Med. 2019;6:65. doi:10.3389/fcvm.2019.00065
- Qi H, Liu Y, Li S, et al. Activation of AMPK Attenuated Cardiac Fibrosis by Inhibiting CDK2 via p21/p27 and miR-29 Family Pathways in Rats. Mol Ther Nucleic Acids. 2017;8:277–290.
- Shapiro SD, Ranjan AK, Kawase Y, et al. Cyclin A2 induces cardiac regeneration after myocardial infarction through cytokinesis of adult cardiomyocytes. Sci Transl Med. 2014;6(224):224ra27. doi:10.1126/scitranslmed.3007668
- Liu P, Sun H, Zhou X, et al. CXCL12/CXCR4 axis as a key mediator in atrial fibrillation via bioinformatics analysis and functional identification. Cell Death Dis. 2021;12(9):813. doi:10.1038/s41419-021-04109-5
- Chu PY, Joshi MS, Horlock D, Kiriazis H, Kaye DM. CXCR4 Antagonism Reduces Cardiac Fibrosis and Improves Cardiac Performance in Dilated Cardiomyopathy. Front Pharmacol. 2019;10:117. doi:10.3389/fphar.2019.00117