Abstract
Background
Efficient target-specific siRNA delivery has always been a primary concern in the field of siRNA clinical application.
Purpose
In this study, four different types of anti-epidermal growth factor receptor (EGFR) antibody-conjugated immunonanoparticles were prepared and tested for cancer cell-targeted therapeutic siRNA delivery.
Materials and methods
The prepared nanoparticles encapsulating siRNAs were character-ized by gel retardation and particle analysis using a Zetasizer. In vitro transfection and reduction of target genes, vimentin and JAK3, were determined using quantitative reverse transcription polymerase chain reaction. In vivo tumor targeting and antitumoral efficacies of the nanoparticles were evaluated in mice carrying tumors.
Results
Among these immunonanoparticles, anti-EGFR immunolipoplexes and immunoviroplexes exhibited remarkable cell binding and siRNA delivery to EGFR-expressing tumor cells compared to immunoliposomes and immunovirosomes. Especially, the anti-EGFR immunoviroplexes exhibited the most efficient siRNA transfection to target tumor cells. Therefore, antitumoral vimentin and Janus kinase-3 siRNAs were loaded in the anti-EGFR immunolipoplexes and immunoviroplexes, which were tested in mice carrying SK-OV-3 tumor xenografts. In fact, the therapeutic siRNAs were efficiently delivered to the tumor tissues by both delivery vehicles, resulting in significant inhibition of tumor growth. Moreover, administration of doxorubicin in combination with anti-EGFR immunoviroplexes resulted in remarkable and synergistic tumor growth inhibition.
Conclusion
This study provides experimental proof that cancer cell-targeted immunoviroplexes are an efficient siRNA delivery system for cancer therapy. Moreover, this study also suggests that a combination of conventional chemotherapy and tumor-directed anticancer siRNA therapy would be a better modality for cancer treatment.
Introduction
RNA interference (RNAi) has gained much attention since 1998 because of its pivotal role in gene expression and regulation.Citation1 Gene therapy using siRNA represents a potent and specific method of targeted gene therapy.Citation2,Citation3 A number of siRNA molecules have been suggested as functional mediators of RNAi that can achieve sequence-specific gene knockdown in cancer cells.Citation4,Citation5 Despite the powerful therapeutic potential of siRNA as an anticancer drug, it must be delivered within the target cell to exploit its therapeutic activity.
Therefore, a major challenge in siRNA-based therapy is developing an efficient systemic delivery system. Although they require further improvement, liposomal delivery systems have been successfully utilized in the systemic delivery of siRNAs.Citation6 Lipoplexes, complexes of cationic liposomes and nucleic acids, have been widely utilized for transfection in vitroCitation7 but not in vivo. Previously, lipid nano-particles consisting of cationic lipids and polymers, coupled with tumor-targeting ligands, effectively delivered siRNAs into target tumor cells.Citation8,Citation9 In addition, the development of virosomes containing fusogenic viral envelop proteins has been proven to be more effective in gene transfer than conventional lipoplex-es.Citation10 However, a great deal of effort is still required for efficient siRNA delivery to target cells for clinical applications.
Target-specific delivery has always been a primary concern in the field of clinical application of therapeutic siRNAs.Citation11,Citation12 Various monoclonal antibodies (mAbs) or mAb fragments against certain tumor antigens have been used as ligands for tumor-targeted delivery of siRNA.Citation13 We have also shown that coupling of cetuximab, an antibody (Ab) against epidermal growth factor receptor (EGFR), to the liposomal surface enhances siRNA delivery to target tumor cells in vivo. Over-expression of EGFR has been linked with tumor malignancy, including tumor invasion, angiogenesis, and metastasis.Citation14–Citation16 Elevated levels of EGFR are found in various types of solid tumors, such as breast, ovarian, lung, head and neck, prostate, and colorectal cancers. EGFR overexpression has been considered as a prognostic marker for cancer progression and drug resistance in chemotherapy.Citation17–Citation20 Therefore, EGFR has been recognized as a therapeutic target for cancer treatment.
As far as anticancer siRNAs are concerned, a number of cellular proteins essential in maintaining cell viability have been targeted. In this study, we tested a variety of siRNA molecules to identify the best candidate for anticancer therapy. Among the tested molecules, vimentin (Vim), a member of the type III intermediate filaments, is an important structural protein in adhesion, migration, survival, and various cell signaling processes in activated cells.Citation21 Vim deficiency results in delayed wound healing and reduced cell viability.Citation22 Therefore, the antitumoral therapeutic efficacy of Vim siRNA has been evaluated in several in vitro and in vivo systems.Citation23,Citation24 Janus kinases (JAKs) are also known to be essential for cell proliferation and survival.Citation25 Among them, the activating mutations in JAK3 have been identified as a cause of cancer,Citation26 and therefore, JAK3-targeted gene therapy has been carefully tested for cancer treatment.Citation27
Based on recent reports, we proposed that a therapeutic strategy involving the use of innovative tumor-targeted immu-nonanoparticles for siRNA transfection of tumor cells could be a plausible option for anticancer gene therapy. The present study is designed to develop efficient anti-EGFR immunonano-particles for targeted siRNA delivery to EGFR-expressing cancer cells. The antitumoral activities of Vim and JAK3 siRNAs delivered by anti-EGFR immunonanoparticles were also investigated in mice carrying tumor xenografts. This study provides useful insights into the alternative delivery systems for cancer cell-targeted siRNA delivery.
Materials and methods
Materials
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (DSPE-mPEG2000), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (DSPE-PEG2000-Mal), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Rho-DOPE) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL, USA). O,O′-dimyristyl-N-lysyl glutamate (DMKE) cationic lipid was provided by KOMA Biotech (Seoul, Korea).
The FITC-labeled control siRNA (SN-1023) was purchased from Bioneer (Daejeon, Korea). Luciferase siRNA (GP-RNA1041) and Vim siRNA (GP-RNA1015) were purchased from Genolution Pharmaceuticals, Inc. (Seoul, Korea). JAK3 siRNA (VHS41248) was purchased from Thermo Fisher Scientific (Waltham, MA, USA).
Cells and cell culture
The SK-OV-3 human ovarian carcinoma cell line was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 medium (Thermo Fisher Scientific). B16BL6 (mouse melanoma) cells were provided by Korean Cell Line Bank (Seoul, Korea). They were maintained in minimum essential medium (MEM; Thermo Fisher Scientific). The culture media were supplemented with 10% heat-inactivated fetal bovine serum (FBS; Thermo Fisher Scientific), 100 units/mL penicillin, and 100 μg/mL streptomycin (Thermo Fisher Scientific). The cells were grown in a humidified atmosphere of 95% air and 5% CO2 at 37°C.
Purification of Sendai virus F/HN protein
As described previously,Citation28 Sendai viral F/HN proteins were purified from the cultured Sendai virus (ATCC No 1698936, VR 907). Briefly, the Sendai virus was cultured in the allantoic fluid of egg embryo and then was centrifuged at 100,000×g for 1 hour at 4°C. The pellet was resuspended in 2 mL of phosphate buffered saline (PBS) containing 1% Triton X-100. After incubation for 2 hours at 20°C, the suspension was centrifuged at 100,000×g for 1 hour at 4°C and the detergent was then removed from the supernatant containing F/HN proteins by stepwise addition of Bio-Beads SM-2 (Bio-Rad Laboratories Inc., Hercules, CA, USA) with constant rocking.
Preparation of immunonanoparticles
Preparation of liposomes and virosomes encapsulating siRNA
DMKE (5 mole%), POPC (91 mole%), DSPE-mPEG2000 (3.8 mole%), and DSPE-PEG2000-Mal (0.2 mole%) were dissolved in a mixture of chloroform and methanol (2:1, v/v). The organic solvent was completely evaporated, and the dried lipid films (2 mg lipids) were then hydrated in 1 mL of 0.1 M phosphate buffer (pH 5.5) containing siRNA (10:1, a weight ratio of lipid and siRNA). After hydration, the liposomes were subjected to 10 cycles of freezing and thawing and extruded 10 times through a polycarbonate membrane with a pore size ranging from 80 to 800 nm using an extruder (Avanti Polar Lipids, Inc.). For virosome preparation, the F/HN protein solution was added to the liposome solution (10:1, a weight ratio of lipid and F/HN protein) and then incubated for 15 minutes at room temperature with gentle mixing. The preparation buffer was changed to phosphate buffer (pH 7.2, 0.1 M).
Preparation of cationic lipoplexes and cationic viroplexes
DMKE (48 mole%), cholesterol ([chol] 48 mole%), DSPE-mPEG2000 (3.8 mole%), and DSPE-PEG2000-Mal (0.2 mole%) were dissolved in the organic solvent. DMKE cationic liposomes were prepared as described earlier in phosphate buffer (pH 7.2, 0.1 M). For effective siRNA complexation, siRNA molecules were precomplexed with protamine sulfate (PS; Sigma-Aldrich Co., St Louis, MO, USA; 0.35:1, a weight ratio of siRNA and PS) for 30 minutes at room temperature. Lipoplexes were prepared by gentle mixing of PS-condensed siRNA and cationic liposomes at various N/P ratios. For viroplex preparation, as described earlier, the F/HN protein solution was added to the lipoplex solution (10:1, a weight ratio of lipid and F/HN protein) and then additionally incubated for 15 minutes at room temperature with gentle mixing.
Conjugation of anti-EGFR Ab to nanoparticles
Anti-EGFR Ab (cetuximab; BMS, New York, NY, USA) was thiolated for 1 hour at room temperature by Traut’s reagent in phosphate buffer (0.1 M, 2 mM EDTA, pH 8.0). Unreacted Traut’s reagent was removed by passing through PD-10 column. The thiolated Ab solution was added to the prepared solution of lipid nanoparticles (0.2:1, a molar ratio of Ab and maleimide) and incubated for 20 hours at 4°C with continuous shaking. Unconjugated Abs were removed by passing through a Sepharose CL-4B column in phosphate buffer (pH 7.2, 0.1 M).
Gel retardation analysis of anti-EGFR immunonanoparticles containing siRNA
To ensure effective siRNA encapsulation (or complexation), the immunonanoparticles prepared under varied conditions ran on 1% agarose gels. An appropriate N/P ratio of the vehicles and the amount of encapsulated siRNA were ana-lyzed by the Quantity One program of Gel Doc EZ system (Bio-Rad Laboratories Inc.).
Analysis of vesicle size and surface charge
To observe the changes in vesicular size and surface charge of the anti-EGFR immunonanoparticles during siRNA encapsulation (or complexation) and Ab coupling, the particle size and ζ-potential were measured using Zetamaster S (Malvern instruments, Malvern, UK).
In vitro cell binding and transfection by anti-EGFR immunonanoparticles
Specific cellular binding of the vehicles was assayed with rhodamine-labeled anti-EGFR immunonanoparticles containing FITC-labeled siRNA. EGFR-expressing SK-OV-3 cells and B16BL6 cells expressing no EGFR (4×105 cells/well) were treated with the anti-EGFR immunonanoparticles in six-well plates (1 μg of siRNA per well) for 30 minutes at room temperature. In order to verify the EGFR-mediated siRNA transfection, cells in another set were pretreated with free cetuximab (1 μg/mL per well) for 30 minutes at room temperature. The cells were then treated with 2% paraformaldehyde for 5 minutes at room temperature in the dark. The fixed cells were counted using a flow cytometer (Becton Dickinson, San Jose, CA, USA).
To verify their transfection capabilities, the same cancer cells were pre-transfected with cationic DMKE/Chol lipoplexes of luciferase-encoding plasmid pAAVCMV-Luc (1 μg of pDNA) 24 hours earlier, and then treated with the immunonanoparticles prepared under varied conditions (20 nM luciferase siRNA). After transfection in the absence of serum for 4 hours at 37°C, the cells were additionally incubated in 10% FBS-containing media for 24 hours at 37°C. The transfected cells were lysed with 200 μL of lysis buffer (1% Triton X-100, 1 mM dithiothreitol, and 2 mM EDTA, pH 7.8) for 2 hours at room temperature with gentle agitation. The culture plates were stored at −20°C for 20 minutes and thawed at room temperature. The cell lysates were centrifuged at 12,000×g for 20 minutes at 4°C. Luciferase activities in the supernatants were measured with a luciferase assay kit (Thermo Fisher Scientific) and a luminometer (Minilumat LB9506; Berthold Technologies, Bad Wildbad, Germany). The data were expressed as relative light unit (RLU) of luciferase per milligram of total cellular proteins.
Cell cytotoxicity of anti-EGFR immunonanoparticles containing anticancer siRNA
Cell cytotoxicity was measured using Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan). SK-OV-3 cells were seeded into 96-well plates (5×103 cells/well) and cultured for 24 hours. The tumor cells were treated with Vim and/or JAK3 siRNA (20 nM) encapsulated in anti-EGFR immunonano-particles and then further cultured for 48 hours. Following this, 10 μL of tetrazolium substrate was added to each well and further incubated at 37°C for 2 hours. The absorbance of each well was measured at 450 nm (n=5).
Quantitative reverse transcription polymerase chain reaction (RT-qPCR) analysis
The prepared anti-EGFR immunonanoparticles containing Vim or JAK3 siRNA were added to SK-OV-3 and B16BL6 cells (4×105 cells/well, 20 nM siRNA). After transfection (as described earlier), the cells were washed twice with PBS (pH 7.4) and treated with TRIzol reagent (Thermo Fisher Scientific) to extract total RNA. Complementary DNA (cDNA) was synthesized using a Reverse Transcription Master Premix (ELPIS, Daejeon, Korea) according to the manufacturer’s protocol. The total RNA was added to the master mix, incubated at 65°C for 5 minutes and then quickly chilled on ice. cDNA synthesis reaction was performed at 25°C for 10 minutes, 37°C for 50 minutes, and 70°C for 15 minutes.
The relative mRNA expression levels of Vim and JAK3 were measured by a RT-qPCR employing SYBR Green using a CFX-96 real-time polymerase chain reaction (PCR) system (Bio-Rad Laboratories Inc.). The RT-qPCR was carried out using Thunderbird SYBR qPCR Mix (Toyobo, Osaka, Japan) according to the manufacturer’s protocols. Real-time PCR amplification for Vim and JAK3 mRNA was performed under the following conditions: 95°C for 3 minutes, followed first by 10 cycles of 15 seconds at 95°C and 30 seconds at 60°C and then 40 cycles of 15 seconds at 95°C and 30 seconds at 55°C. The mRNA expression level was quantified by determining the cycle threshold (CT), which is the number of PCR cycles required for the fluorescence to exceed a value significantly higher than the background fluorescence. To avoid false negatives due to degradation of mRNA, a GAPDH reference gene was used as a control. The forward and reverse PCR primers (XenoTech, Daejeon, Korea) used were as follows: Vim, forward: 5′-TCACTCCCTCTGGTTGATAC-3′ and reverse: 5′-GTCATCGTGATGCTGAGAAG-3′; JAK3, forward: 5′-TGAGAAGGCTCAGAGTAAGG-3′ and reverse: 5′-CCAGAGAGGGAAAGGAACT-3′; and GAPDH, forward: 5′-CCATCTTCCAGGAGCGAGATCC-3′ and reverse: 5′-ATGGTGGTGAAGACGCCAGTG-3′.
Animal studies
Animal protocols were approved by the Yonsei University at Wonju Animal Care and Use Committee in accordance with the regulations of the Association for Assessment and Accreditation of Laboratory Animal Care International. Female BALB/c nude mice (4–5-week old; Orient Bio, Seongnam, Korea) were subcutaneously injected with 1×107 of SK-OV-3 cells in the right abdomen.
To verify tumor-targeted localization, the anti-EGFR immu-nonanoparticles containing rhodamine-DOPE (0.1 mole%) were intravenously injected to BALB/c nude mice carrying SK-OV-3 tumors. The tumor tissues were excised 24 hours post administration. The excised tumors were immediately frozen and transversally sectioned (3 μm). The sections were then observed using fluorescence microscopy (×100).
The antitumoral therapeutic efficacy of anti-EGFR immunonanoparticles containing anticancer siRNAs was also tested in the same animal tumor model. When SK-OV-3 tumors grew to ~50 mm3 in volume (length×width2/2), the mice (n=5) were intravenously injected with anti-EGFR immunonanoparticles containing Vim and/or JAK3 siRNAs (0.5 mg/kg per mouse). The anti-EGFR immunona noparticles were intravenously administered four times every 3 days. In another set of animal experiment, doxorubicin (6 mg/kg) was intravenously co-injected with the anti-EGFR immunonanoparticles containing Vim and/or JAK3 siRNAs at the same injection days. Tumor growth was monitored for 28 days post treatment. Mice were sacrificed on day 43, and lung colonization was evaluated.
Statistical analyses
Each value was expressed as the mean±SD of multiple test results. The significance of mean value difference was evaluated using Student’s t-test. Differences were considered as significant when the P-value was <0.05.
Results and discussion
Preparation and characterization of anti-EGFR immunonanoparticles containing siRNA
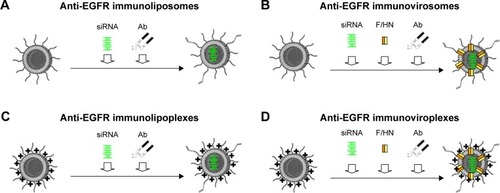
In this study, four different types of liposomes containing siRNA were prepared and their capability to deliver siRNA was evaluated. First, neutral POPC-based liposomes were prepared (). A virosomal formulation of the POPC liposomes was then prepared by reconstitution of Sendai F/HN proteins (). Second, cationic DMKE-based lipoplexes were prepared (). DMKE viroplexes were also prepared by addition of Sendai F/HN proteins to the cationic lipoplexes (). Finally, the prepared lipid nanoparticles were functionalized by conjugation of anti-EGFR Ab molecules to their surface and were referred to as immunonanoparticles.
Figure 1 Schematic illustration of anti-EGFR immunonanoparticles with siRNA.
Notes: Immunoliposomes were prepared by coupling of thiolated anti-EGFR Abs to the surface of neutrally charged liposomes encapsulating siRNA (A). Immunovirosomes were prepared by insertion of Sendai viral F/HN proteins into the liposomes encapsulating siRNA followed by Ab coupling (B). Immunolipoplexes were prepared by Ab coupling to cationic lipoplexes of siRNA (C). Immunoviroplexes were prepared by insertion of Sendai viral F/HN proteins into the lipoplexes of siRNA followed by Ab coupling (D).
Abbreviations: EGFR, epidermal growth factor receptor; Ab, antibody.
Clinically approved cetuximab can serve as a tumor-targeting ligand for EGFR-overexpressing cancer cells. The Ab molecules were conjugated to the surface of the prepared lipid-based siRNA carriers by direct coupling. Thiolated Abs were effectively conjugated to reactive maleimide groups at the PEG termini exposed on their surface. According to gel filtration analysis of the reaction mixtures, the Ab conjugation reaction was almost completed at a 1:5 molar ratio of Ab and DSPE-PEG2000-Mal ().
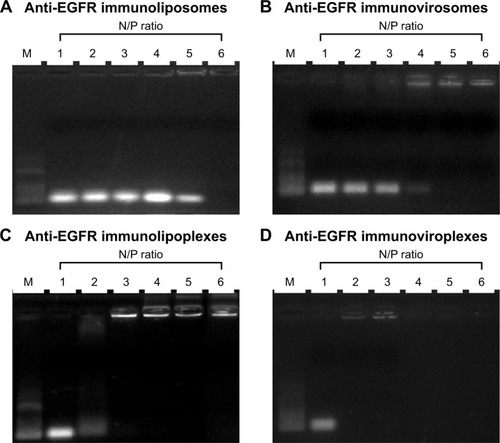
In order to determine an optimal lipid:siRNA ratio for efficient siRNA encapsulation in the vehicles, anti-EGFR immunonanoparticles were hydrated at 1:1–2:1 N/P ratios of cationic lipid and siRNA. The prepared immunonanoparticles were then loaded on 1% agarose gels (). According to gel retardation analysis, encapsulation of siRNA into the anti-EGFR immunoliposomes and immunovirosomes was completed at 12:1 and 9:1 N/P ratios, respectively (). Meanwhile, cationic immunolipoplexes and immunoviroplexes showed completion of siRNA complexation at 6:1 and 3:1 N/P ratios, respectively (). These results showed that the DMKE cationic formulations, including lipoplexes and viroplexes were more efficient in terms of siRNA loading into lipid vesicles. Presumably, charge interaction between the cationic vesicle surface and anionic siRNA molecule resulted in their spontaneous complexation. In addition, F/HN proteins were helpful in siRNA complexation with the cationic vesicles.
Figure 2 Encapsulation or complexation of siRNA with anti-EGFR immunonanoparticles.
Notes: The anti-EGFR immunoliposomes (A), immunovirosomes (B), immunolipoplexes (C), and immunoviroplexes (D) prepared at various N/P ratios of siRNA and DMKE were run on 1% agarose gels and visualized by ultraviolet illumination. Lane M: 100 bp molecular weight markers.
Abbreviations: EGFR, epidermal growth factor receptor; DMKE, O, O′-dimyristyl-N-lysyl glutamate.
DMKE-based cationic vesicles exhibited a more compact size than POPC-based liposomal vesicles (). The lipo-somal formulations with siRNA exhibited a slightly negative surface charge, while the lipoplex formulations were positively charged even after completion of siRNA complexation. As previously shown, addition of F/HN proteins increases the vesicular size of immunonanoparticles. All the prepared anti-EGFR immunonanoparticles had appropriate vesicular size for in vivo systemic applications. Cell toxicity of all the immunonanoparticles was insignificant in the tested concentrations (). All anti-EGFR immunonanoparticle carriers were nearly toxic to EGFR-expressing SK-OV-3 cells or control B16BL6 cells at a concentration of 10 μg lipid/mL.
Table 1 Vesicular size and surface charge of anti-EGFR immu-nonanoparticles containing siRNA
Target cell-specific binding of anti-EGFR immunonanoparticles containing siRNA
To verify the target cell-specific binding of immunonanoparticles, two different cell types, EGFR-positive SK-OV-3 and EGFR-negative B16BL6 cells, were treated with the prepared anti-EGFR immunonanoparticles containing FITC-siRNA. Their cellular bindings were then analyzed by flow cytometry (). As expected, regardless of the nanoparticle type, all anti-EGFR immunonanoparticles showed high cellular binding to EGFR-positive SK-OV-3 cells, comparable to the non-specific cell binding of conventional cationic DMKE liposomes (EzWay™ transfection; KOMA BIOTECH, Seoul, Korea; ). However, the same nanoparticles showed far less binding to EGFR-negative B16BL6 cells ().
Figure 3 Tumor-targeted cell binding and siRNA transfection by anti-EGFR immunonanoparticles.
Notes: SK-OV-3 (A) and B16BL6 (B) cells were incubated with anti-EGFR immunonanoparticles containing FITC-siRNA in the presence or absence of free cetuximab for 30 minutes at 4°C. Binding of immunonanoparticles to the cells was analyzed by flow cytometry. Each bar represents the mean±SD of three independent experiments. In another set of experiment, the same cancer cell lines were pre-transfected with plasmid encoding luciferase (pLuc) in 24-well plates, and 24 hours later, they were transfected with luciferase siRNA by anti-EGFR immunonanoparticles in the presence or absence of free cetuximab for 24 hours (C and D). Luciferase expression in the transfected cells is expressed as RLU per milligram of proteins. Each bar represents the mean±SD of three independent experiments. *P<0.05 and **P<0.01 between test groups.
Abbreviations: EGFR, epidermal growth factor receptor; MFI, mean fluorescence intensity; DMKE, O,O′-dimyristyl-N-lysyl glutamate; Ab, antibody; Chol, cholesterol.
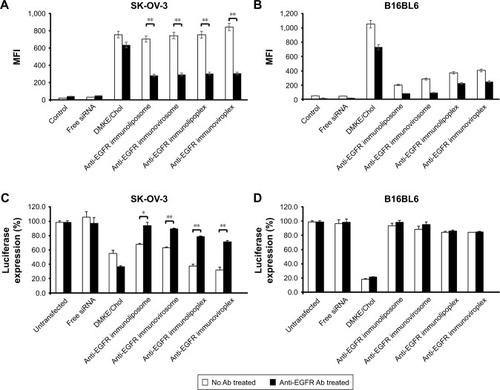
In order to verify whether the effective binding of immunonanoparticles was mediated by EGFR, cancer cells were pretreated with anti-EGFR Ab and then treated with the immunonanoparticles. The cellular binding of conventional cationic lipoplexes was not interfered by addition of neutralizing Ab, but binding of anti-EGFR immunonanoparticles to target cells (SK-OV-3) was significantly inhibited by Ab pretreatment. These results clearly imply that the higher binding efficiency of immunonanoparticles to the EGFR-positive cancer cells is due to specific interactions between cetuximab on the nanoparticle surface and EGFRs overexpressed on the cancer cell surface.
Target-specific transfection of siRNA by anti-EGFR immunonanoparticles
Luciferase-expressing SK-OV-3 and B16BL6 cells were treated with anti-EGFR immunonanoparticles containing luciferase siRNA. Overall, there was a significant reduction of luciferase expression in EGFR-positive SK-OV-3 cells by luciferase siRNA transfection (). Most of all, the reduction of luciferase expression by transfection with immunolipoplexes or immunoviroplexes was higher than that by transfection with immunoliposomes or immunoviro-somes and even higher than that by transfection with DMKE cationic lipoplexes. However, as shown earlier, in EGFR-mediated cell binding, pretreatment of SK-OV-3 cells with free cetuximab significantly interfered with the reduction of luciferase expression. Meanwhile, B16BL6 cells were effectively transfected by DMKE cationic lipoplexes, but not by the immunonanoparticles regardless of the Ab treatment (). These results suggest that anti-EGFR immuno-lipoplexes and immunoviroplexes serve as efficient siRNA delivery systems for targeting cancer cells overexpressing EGFR but result in nonspecific and untargeted transfection in B16BL6 cells. The delivered siRNA molecules were readily translocated into the cytosol and interfered with the expression of target luciferase gene.
In vitro delivery of anticancer siRNA by anti-EGFR immunonanoparticles
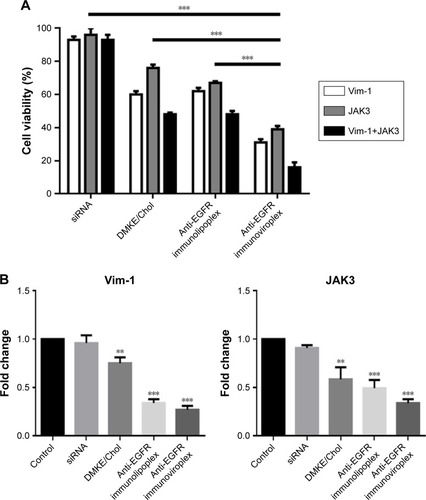
Based on the results of cell binding and siRNA transfection, SK-OV-3 cells were selected for subsequent experiments of anticancer siRNA transfection. To verify the anticancer efficacy of siRNA, SK-OV-3 cells were treated with anti-EGFR immunonanoparticles containing Vim and/or JAK3 siRNAs, and cell toxicity was then measured by the MTT assay. When the cancer cells were transfected with either Vim or JAK3 siRNA using anti-EGFR lipoplexes or viroplexes, their viability was seriously reduced (). Combined treatment with Vim and JAK3 siRNAs showed further inhibition of SK-OV-3 cell growth. This suggests that two different types of siRNA, which have different anticancer mechanisms, may function synergistically (or additively).
Figure 4 In vitro anticancer activity of Vim and JAK3 siRNAs.
Notes: (A) Cell viability was measured by CCK-8 assay 24 hours later. SK-OV-3 cells were treated with anti-EGFR immunonanoparticles containing Vim-1 and/or JAK3 siRNAs. ***P<0.001 between test groups. (B) Relative mRNA expressions of the Vim-1 and JAK3 were analyzed by real-time RT-PCR. The expressions of Vim-1 and JAK3 in SK-OV-3 cells treated with various liposomal formulations were examined 16 hours post transfection using RT-qPCR. The ΔΔCT method was used to calculate the RT-qPCR fold changes using GAPDH mRNA for normalization, and all the expression changes were relative to the untreated control. Triplicate independent RT-qPCR was performed. **P<0.01 and ***P<0.001 vs siRNA-treated.
Abbreviations: Vim, vimentin; CCK-8, Cell Counting Kit-8; RT-PCR, reverse transcription polymerase chain reaction; RT-qPCR, reverse transcription quantitative polymerase chain reaction; DMKE, O,O′-dimyristyl-N-lysyl glutamate.
It has been well established that Vim is the most widely expressed intermediate filament protein and is essential in maintaining cell adhesion, migration, and survival.Citation29–Citation33 In addition, degradation of Vim has been related to a proapoptotic process that can markedly enhance cell death.Citation34 It is also known that JAK-mediated signaling is important for cell differentiation, proliferation, and survival in mammalian cells.Citation25 Dysregulation of JAK3 has been associated with oncogenesis in malignancies, such as hematological and breast cancers.Citation26 Therefore, inhibition of JAK3 is a promising strategy for cancer gene therapy.Citation27,Citation35
In order to verify the cell death resulting from interference of target genes, the mRNA levels of Vim and JAK3 were determined 16 hours post transfection by RT-qPCR. The mRNA levels of Vim and JAK3 in the transfected SK-OV-3 cells were significantly interfered by transfection with the corresponding siRNAs loaded in the anti-EGFR immunoviroplexes and immunoviroplexes (). The reduction in mRNA levels by the immunonanoparticles was greater than that by conventional DMKE lipoplexes, a potent transfection system. These results imply that anti-EGFR immunolipoplexes and immunoviroplexes are efficient siRNA delivery systems for targeting EGFR-overexpressing tumor cells in vitro.
Intratumoral localization of anti-EGFR immunonanoparticles containing siRNA
To verify the tumor targetability of the anti-EGFR immu-nonanoparticles, rhodamine-labeled anti-EGFR immu-nolipoplexes and immunoviroplexes were intravenously injected to BALB/c nude mice carrying SK-OV-3 tumors. Cryosections of SK-OV-3 xenografts were then observed by fluorescence microscopy (). Both the tumor-targeted immunonanoparticles showed more effective localization in the tumor tissues compared to conventional DMKE/Chol lipoplexes.
Figure 5 In vivo localization of anti-EGFR immunonanoparticles in tumors.
Notes: Rhodamine-labeled anti-EGFR immunonanoparticles were intravenously administered to mice carrying SK-OV-3 tumors. In all, 24 hours post injection, tumor tissues were dissected and then examined using a fluorescence microscope (×100).
Abbreviations: EGFR, epidermal growth factor receptor; DMKE, O,O′-dimyristyl-N-lysyl glutamate; Chol, cholesterol.
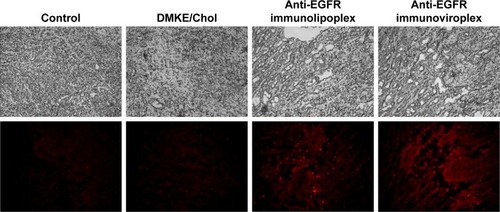
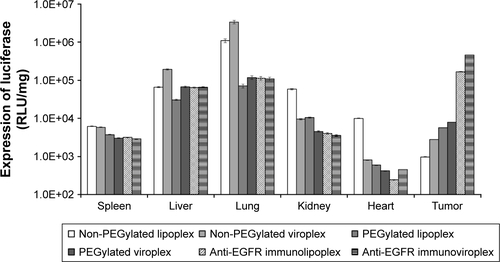
The administered immunonanoparticles were cationi-cally charged on their surface (). Nevertheless, the PEGylated immunonanoparticles were able to efficiently target the SK-OV-3 tumor tissues. This implies that PEGylation of cationic immunolipoplexes and immunoviroplexes with 4 mole% of DSPE-mPEG2000 may provide adequate steric hindrance from serum components, resulting in less transgene expression in the first capillary bed of pulmonary vasculature and the reticuloendothelial system (RES) in the liver and spleen (). These results indicate that PEGylation of lipid nanoparticles presumably provide further stability in maintaining nanoparticle membrane integrity, resulting in less accumulation in the lungs. The fully PEGylated immuno-lipoplexes and immunoviroplexes were able to circulate long enough to be extravasated and then recognized by EGFRs on the tumor cell surface.
In vivo antitumoral activity of anti-EGFR immunonanoparticles containing Vim and JAK3 siRNAs
In order to examine the antitumoral activity of Vim and JAK3 siRNAs combined, anti-EGFR immunonanoparticles containing Vim and/or JAK3 siRNA were prepared and intravenously administered to mice carrying SK-OV-3 tumors four times at intervals of 3 days. During the 28 days of treatment, mice treated with both Vim and JAK3 siRNAs encapsulated in immunoliposomes or immunovirosomes exhibited slower tumor growth than those treated with either one of the siRNAs alone (). Under the same experimental conditions, virosomal formulations were generally more effective in inhibiting SK-OV-3 cancer progression. As expected, pulmonary metastasis of SK-OV-3 xenografts was proportionally correlated with tumor growth. Mice co-transfected with the two different siRNAs had a smaller number of tumor colonies in the lungs compared to those treated with either one of the siRNAs alone (). These results highlighted two important points in in vivo transfection of anticancer therapeutic siRNAs. First, the prepared anti-EGFR immu-nolipoplexes and immunoviroplexes were able to deliver siRNA molecules efficiently to the tumor tissues, resulting in significant inhibition of tumor progression. Second, in vivo co-transfection of Vim and JAK3 siRNAs was more effective in inhibition of tumor progression compared to transfection with either one of the siRNAs alone, as shown earlier (). Recently, there have been a few reports regarding enhanced antitumoral effects of combinatorial RNAi therapy. For example, intravenous administration of immunolipoplexes with EGFR siRNA and telomerase reverse transcriptase siRNA showed an additive inhibitory effect on tumor progression in mice.Citation36
Figure 6 Inhibition of tumor progression by intravenous administration of anti-EGFR immunonanoparticles containing Vim-1 and/or JAK3 siRNAs.
Notes: Anti-EGFR immunonanoparticles containing Vim-1 and/or JAK3 siRNAs were intravenously administered to mice carrying SK-OV-3 tumors. The tumor growth was measured for 25 days (E) or 28 days (A and C), and the pulmonary tumor colonies (B, D, and F) were counted on the 28th day after the treatment. First, the mice were injected with immunonanoparticles containing Vim-1, JAK3, or Vim-1 and JAK3 siRNAs (A and B). Second, EGFR-targeted or -untargeted nanoparticles containing Vim-1 and JAK3 siRNAs were injected into the mice (C and D). Lastly, a combinatorial treatment of doxorubicin and immunonanoparticles with Vim-1 and JAK3 was given to the mice (E and F). The data represent the mean±SD of n=5. *P<0.05 and **P<0.01 between test groups.
Abbreviation: Vim, vimentin.
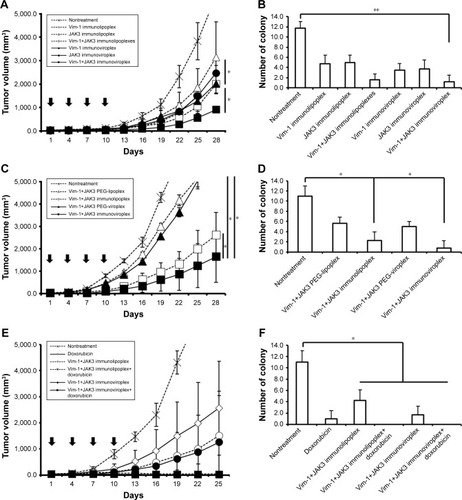
In another set of mouse experiment, the EGFR-targeted and -untargeted nanoparticles were compared in terms of tumor growth and pulmonary colonization. The anti-EGFR immunoviroplexes and immunolipoplexes containing both Vim and JAK3 siRNAs were intravenously administered, and their antitumoral effects were then compared with that of untargeted nanoparticles (). Anti-EGFR immunoviroplexes had superior antitumoral effects compared to immunolipoplex formulations, as shown earlier. More importantly, compared to the untargeted formulation, both EGFR-targeted formulations resulted in ~75%–84% reduction of tumor growth and further decrease in lung colonization on day 28 after the first treatment.
Combinatorial treatment with Vim/JAK3 siRNA and doxorubicin
In the third set of mouse experiment, mice carrying tumors were co-treated with immunonanoparticles containing Vim and JAK3 siRNA and doxorubicin. Mice treated with Vim/JAK3 siRNA in addition to doxorubicin showed least growth of tumor xenografts, regardless of the immunoli-poplexes or immunoviroplexes (). In addition, three out of five mice co-treated with doxorubicin and the therapeutic siRNAs showed complete regression of implanted tumors and no sign of pulmonary colonization. These results suggest that the combinatorial treatment synergistically (or additively) drives their inhibitory actions put together in tumor progression, presumably resulting in effective inhibition of tumor growth and pulmonary metastasis. As mentioned previously, Vim is a critical structural
Conclusion
This study shows that a rationale of targeted delivery of therapeutic siRNAs mediated by specific recognition of EGFRs on the tumor cell surface could be realized. It also suggests that a combination of RNAi therapy and conventional chemotherapy can be considered as a plausible modality for effective cancer treatment. Among the tested anti-EGFR immunonanoparticles, the immunoviroplex formulation was the most efficient in delivering siRNA to target cancer cells, and therefore, it could be widely utilized for future RNAi-based clinical applications.
Acknowledgments
This research was funded by the National Research Foundation of Korea (2013R1A1A2009431, 2015M2B2A4031262) and the National Cancer Center of Korea (09202002_14918).
Supplementary materials
Figure S1 Elution profiles of anti-EGFR immunonanoparticles and unbound Abs.
Notes: The reaction mixtures of nanoparticles (liposomes and virosomes) and cetuximab antibodies (1:5 molar ratio of antibody and DSPE-PEG2000-Mal) were loaded onto a Sepharose CL-4B column and then eluted. The antibody concentration in each eluted fraction (1 mL) was quantified by colorimetric protein assay.
Abbreviations: EGFR, epidermal growth factor receptor; Ab, antibody; DSPE-PEG2000-Mal, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000].
![Figure S1 Elution profiles of anti-EGFR immunonanoparticles and unbound Abs.Notes: The reaction mixtures of nanoparticles (liposomes and virosomes) and cetuximab antibodies (1:5 molar ratio of antibody and DSPE-PEG2000-Mal) were loaded onto a Sepharose CL-4B column and then eluted. The antibody concentration in each eluted fraction (1 mL) was quantified by colorimetric protein assay.Abbreviations: EGFR, epidermal growth factor receptor; Ab, antibody; DSPE-PEG2000-Mal, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000].](/cms/asset/d7926df3-bf1b-469a-bf35-b4e66d8ccdb0/dijn_a_12194045_sf0001_b.jpg)
Figure S2 Cell toxicity of anti-EGFR immunonanoparticles.
Notes: SK-OV-3 (A) and B16BL6 (B) cells were treated with the various concentrations of anti-EGFR immunonanoparticles (immunoliposomes, immunovirosomes, immunolipoplexes, and immunoviroplexes) at 37°C for 24 h. Cell viability was measured by the MTT assay.
Abbreviations: EGFR, epidermal growth factor receptor; DMKE, O,O′-dimyristyl-N-lysyl glutamate.
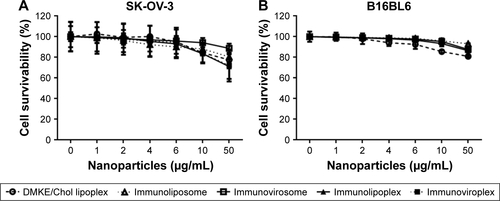
Figure S3 In vivo transfection of luciferase gene by anti-EGFR immunonanoparticles.
Notes: The anti-EGFR immunonanoparticles containing pLuc were intravenously administered to SK-OV-3-xenografted nude mice. Mice were sacrificed 24 hours post injection, and luciferase expression in the spleen, liver, lungs, kidneys, heart, and tumor tissues was measured and expressed as RLU per milligram of protein.
Abbreviations: EGFR, epidermal growth factor receptor; pLuc, plasmid encoding luciferase; RLU, relative light unit.
Disclosure
The authors report no conflicts of interest in this work.
References
- de JongeJHoltropMWilschutJHuckriedeAReconstituted influenza virus envelopes as an efficient carrier system for cellular delivery of small-interfering RNAsGene Ther200613540041116267567
- de FougerollesAVornlocherHPMaraganoreJLiebermanJInterfering with disease: a progress report on siRNA-based therapeuticsNat Rev Drug Discov20076644345317541417
- GaoJLiuWXiaYThe promotion of siRNA delivery to breast cancer overexpressing epidermal growth factor receptor through anti-EGFR antibody conjugation by immunoliposomesBiomaterials201132133459347021296406
- FavaroEIndraccoloSGene therapy of cancer in the clinic: good news in sight from Asia?Curr Opin Mol Ther20079547748217932811
- PirolloKFRaitAZhouQMaterializing the potential of small interfering RNA via a tumor-targeting nanodelivery systemCancer Res20076772938294317409398
- TsengYCMozumdarSHuangLLipid-based systemic delivery of siRNAAdv Drug Deliv Rev200961972173119328215
- LiSHuangLNonviral gene therapy: promises and challengesGene Ther200071313410680013
- LiSDChonoSHuangLEfficient gene silencing in metastatic tumor by siRNA formulated in surface-modified nanoparticlesJ Control Release20081261778418083264
- LiSDChenYCHackettMJHuangLTumor-targeted delivery of siRNA by self-assembled nanoparticlesMol Ther200816116316917923843
- SandersonCMAvalosRKunduANayakDPInteraction of Sendai viral F, HN, and M proteins with host cytoskeletal and lipid components in Sendai virus-infected BHK cellsVirology199520927017077778306
- BakerMRNA interference: homing in on deliveryNature2010464729212251228
- MorilleMPassiraniCVonarbourgAClavreulABenoitJPProgress in developing cationic vectors for non-viral systemic gene therapy against cancerBiomaterials20082924–253477349618499247
- GaoJFengSSGuoYAntibody engineering promotes nanomedicine for cancer treatmentNanomedicine2010581141114521039191
- NicholsonRIGeeJMHarperMEEGFR and cancer prognosisEur J Cancer2001374suppl 4915
- RansonMZD1839 (Iressa): for more than just non-small cell lung cancerOncologist20027suppl 4162412202784
- RaymondEFaivreSArmandJPEpidermal growth factor receptor tyrosine kinase as a target for anticancer therapyDrugs200060suppl 11523 discussion 41–1211129168
- MishaniEHagoolyAStrategies for molecular imaging of epidermal growth factor receptor tyrosine kinase in cancerJ Nucl Med20095081199120219617320
- TolmachevVFriedmanMSandströmMAffibody molecules for epidermal growth factor receptor targeting in vivo: aspects of dimerization and labeling chemistryJ Nucl Med200950227428319164241
- GelovaniJGMolecular imaging of epidermal growth factor receptor expression-activity at the kinase level in tumors with positron emission tomographyCancer Metastasis Rev200827464565318626573
- MishaniEAbourbehGRozenYNovel carbon-11 labeled 4-dimethylamino-but-2-enoic acid [4-(phenylamino)-quinazoline-6-yl]-amides: potential PET bioprobes for molecular imaging of EGFR-positive tumorsNucl Med Biol200431446947615093817
- LahatGZhuQSHuangKLVimentin is a novel anti-cancer therapeutic target; insights from in vitro and in vivo mice xenograft studiesPLoS One201054e1010520419128
- EckesBColucci-GuyonESmolaHImpaired wound healing in embryonic and adult mice lacking vimentinJ Cell Sci2000113pt 132455246210852824
- Bargagna-MohanPHamzaAKimYEThe tumor inhibitor and antiangiogenic agent withaferin A targets the intermediate filament protein vimentinChem Biol200714662363417584610
- McinroyLMäättäADown-regulation of vimentin expression inhibits carcinoma cell migration and adhesionBiochem Biophys Res Commun2007360110911417585878
- WuWSunXHJanus kinase 3: the controller and the controlledActa Biochim Biophys Sin201244318719622130498
- O’SheaJJGadinaMSchreiberRDCytokine signaling in 2002: new surprises in the Jak/Stat pathwayCell2002109supplS121S13111983158
- KlinkMKielbikMNowakMBednarskaKSulowskaZJAK3, STAT3 and CD3-zeta signaling proteins status in regard to the lymphocytes function in patients with ovarian cancerImmunol Invest201241438239822221142
- KimHSKimJSLeeYKKooKHParkYSAn efficient liposomal gene delivery vehicle using Sendai F/HN proteins and protamineCancer Gene Ther200815421422418259213
- LaneEBHoganBLKurkinenMGarrelsJICo-expression of vimentin and cytokeratins in parietal endoderm cells of early mouse embryoNature198330359197017046190091
- HendrixMJSeftorEAChuYWCoexpression of vimentin and keratins by human melanoma tumor cells: correlation with invasive and metastatic potentialJ Natl Cancer Inst19928431651741371813
- O’RourkeNADaileyMESmithSJMcconnellSKDiverse migratory pathways in the developing cerebral cortexScience199225850802993021411527
- HendrixMJSeftorEAChuYWTrevorKTSeftorRERole of intermediate filaments in migration, invasion and metastasisCancer Metastasis Rev19961545075259034607
- WangNStamenovicDMechanics of vimentin intermediate filamentsJ Muscle Res Cell Motil2002235–653554012785103
- ByunYChenFChangRTrivediMGreenKJCrynsVLCaspase cleavage of vimentin disrupts intermediate filaments and promotes apoptosisCell Death Differ20018544345011423904
- KimBHMinYSChoiJSBenzoxathiol derivative BOT-4-one suppresses L540 lymphoma cell survival and proliferation via inhibition of JAK3/STAT3 signalingExp Mol Med201143531332121499010
- HuYShenYJiBWangLZhangZZhangYCombinational RNAi gene therapy of hepatocellular carcinoma by targeting human EGFR and TERTEur J Pharm Sci201142438739121238587
