?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Introduction
The application of nanoparticles (NPs) in medicine and biology has received great interest due to their novel features. However, their adverse effects on the biological system are not well understood.
Materials and methods
This study aims to evaluate the effect of cerium oxide nanoparticles (CNPs) on conformational changes of human hemoglobin (HHb) and lymphocytes by different spectroscopic (intrinsic and synchronous fluorescence spectroscopy and far and near circular dichroism [CD] spectroscopy), docking and cellular (MTT and flow cytometry) investigations.
Results and discussion
Transmission electron microscopy (TEM) showed that CNP diameter is ~30 nm. The infrared spectrum demonstrated a strong band around 783 cm−1 corresponding to the CNP stretching bond. Fluorescence data revealed that the CNP is able to quench the intrinsic fluorescence of HHb through both dynamic and static quenching mechanisms. The binding constant (Kb), number of binding sites (n), and thermodynamic parameters over three different temperatures indicated that hydrophobic interactions might play a considerable role in the interaction of CNPs with HHb. Synchronous fluorescence spectroscopy indicated that microenvironmental changes around Trp and Tyr residues remain almost unchanged. CD studies displayed that the regular secondary structure of HHb had no significant changes; however, the quaternary structure of protein is subjected to marginal structural changes. Docking studies showed the larger CNP cluster is more oriented toward experimental data, compared with smaller counterparts. Cellular assays revealed that CNP, at high concentrations (>50 µg/mL), initiated an antiproliferative response through apoptosis induction on lymphocytes.
Conclusion
The findings may exhibit that, although CNPs did not significantly perturb the native conformation of HHb, they can stimulate some cellular adverse effects at high concentrations that may limit the medicinal and biological application of CNPs. In other words, CNP application in biological systems should be done at low concentrations.
Introduction
Research on the interactions of nanoparticles (NPs) with cells and biomolecules such as protein and DNA has potentially received a great interest in the field of biotechnology and biomedicine for decades. The nature and kind of interactions influence the bio-safety, delivery mechanisms, drug design, and therapeutic efficacy. Therefore, studies on the interaction of nanomaterials with cells and biomolecules can open new avenues in understanding the structural characteristic required for the bio-affinity of NPs toward pharmacological activity.Citation1–Citation4 Nanomaterials can demonstrate a strong influence on cell viability and structural and corresponding functional features of biomolecules. Therefore, continuous interest has been focused toward comprehending the fundamental effects of NPs for interaction with biological systems.Citation5–Citation7
Cerium oxide NPs (CNPs; nanoceria) have shown a number of biomedical features such as antiemetic, bacteriostatic, bactericidal, immunomodulatory, and antitumor activities.Citation8 Moreover, CNPs have been shown to improve mild traumatic brain injury in vitro and in vivo and provide neuroprotective potential for focal cerebral ischemic stroke.Citation9,Citation10 Furthermore, CNPs have stimulated the proliferation of primary mouse embryonic fibroblasts in vitro.Citation11
Recently, CNPs have been shown to have a potential application in the development of catalytic antioxidants in biology and medicine.Citation12,Citation13 CNPs have commonly showed antioxidant activity as biomimetic enzymes and have demonstrated a capacity to actively scavenge a variety of free radicals in vitro and in vivo.Citation12–Citation16
Blood components are known to be one of the first biological systems that interact with nanomaterials, which generates great interest to explore the reaction of different blood components against NPs.
Human hemoglobin (HHb) has a quaternary structure with four subunits.Citation17 Each subunit is composed of a protein chain with a dominant α-helical structure tightly associated with a non-protein prosthetic heme group.Citation17 HHb is known as a two-way respiratory transport protein, carrying oxygen and carbon dioxide between the lungs and tissues.Citation18 In addition, HHb plays an exceptional role in not only a buffering actionCitation18 for oxygen, but also acts as a potential agent in the transport of a number of drugs to their targeted tissues.Citation19,Citation20
To further investigate the side effects of CNPs in the bloodstream, lymphocyte cell viability was explored to examine the effects of CNPs on the basic immunological functions of human lymphocytes.
Optical techniques such as near and far circular dichroism (CD) and fluorescence spectroscopy were employed as practical tools due to their great sensitivity, speed, and simplicity for studying ligand–protein interactions.Citation21–Citation24 A docking study was employed to verify experimental data.
Furthermore, MTT assay and flow cytometry was carried out to explore the antiproliferative effect of CNPs on lymphocytes.
Materials and methods
Materials
The HHb was obtained from Sigma-Aldrich Company (Sigma-Aldrich, St Louis, MO, USA). The concentration was determined spectrophotometrically on the basis of the Bradford assay. CNPs (99.97%, 30 nm) were purchased from US-Nano Company (USA). All other materials were of analytical grade.
Methods
Preparation of Hb and NPs
Defined concentrations of HHb and CNPs were prepared from fresh stock in phosphate buffer (pH 7.4, 10 mM).Citation25 In order to increase the colloidal stability of CNP, the stock suspension was mixed by vortex for 5 min, and sonicated for 30 min using a sonicator probe (Misonix-S3000, USA) at room temperature.
TEM analysis
Image of CNPs were recorded with a TEM (Zeiss, EM10C, Germany) operated at 100 kV. Samples was prepared by sonication for 30 min using a Misonix sonicator (S3000) and by depositing a few droplets of dilute CNP solution on to a carbon film. The CNP diameter was analyzed to reveal the morphology and diameter of the individual CNP.
Infrared spectroscopy
The Fourier transform infrared (FTIR) spectrum of the CNP was recorded in the transmission mode at room temperature using the potassium bromide (KBr) pellet technique. The KBr was dried in a dryer at 200°C for 24 h. 20 mg KBr was then homogenized with CNPs and, thereafter, ground to fine powder. A Perkin-Elmer FTIR (Model 2000) spectrophotometer was employed to determine the spectrum of CNPs mixed with spectrally pure KBr and then pressed to form thin plates of 0.1 cm−1 thickness. The sample was then subjected to FTIR spectroscopic analysis in the wavelength range of 400–4,000 cm−1.
Measurement of fluorescence during the reaction of CNPs with HHb
Fluorescence measurements were performed using a Hitachi spectrofluorometer (MPF-4 model, Japan). HHb (2.5 µM) was incubated with different concentrations of CNPs (2.5–27.5 µM) in 10 mM potassium phosphate buffer, at pH 7.4, in a total volume of 0.5 mL at different temperatures of 298, 310, and 315 K for 2 min. With an excitation wavelength of 280 nm, two emission wavelengths at 340 and 380 nm for HHb and CNP, respectively, were detected. The slit widths of excitation and emission were set at 5 and 10 nm, respectively. All measurements were corrected against buffer solution and inner filter effect.
Synchronous fluorescence spectra
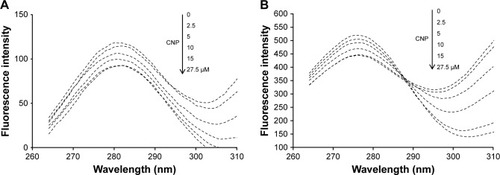
The synchronous fluorescence spectra of protein (2.5 µM) in the presence of CNPs (2.5–27.5 µM) were scanned from 260 to 310 nm with an excitation wavelength of Δλ = 20 (detecting microenvironmental changes of Tyr residue) and Δλ = 60 nm (detecting microenvironmental changes of Trp residue). All measurements were undertaken at room temperature and, finally, were subtracted from the buffer solution and inner filter effect.
CD measurements
CD spectra were recorded using an Aviv model 215 spectropolarimeter (Lakewood, NJ, USA) at room temperature. Changes in the secondary and quaternary structures of HHb in the absence and presence of different concentrations of CNPs were monitored in the far UV-CD region (200–250 nm) and near UV-CD region (260–350 nm), respectively. The protein concentration for far and near CD was 3 and 15 µM, respectively. The concentrations of CNPs for far CD were 3, 10, and 30 µM, whereas the concentrations of CNPs for near CD were 15, 30, and 45 µM. The results are expressed in molar ellipticity [θ] (cm2 dmol−1) with a mean amino acid residue weight of 114 (MRW) for HHb. All CD bands were corrected against buffer and NP solutions.
Molecular docking
To model the CNPs, we used a Ce44O88 cluster as the model of larger particles and a Ce19O32 cluster as the model of smaller particles. Cartesian coordinates of atoms were directly obtained from supplementary files of Loschen et al’s work,Citation25 and these coordinates were converted to pdb files. The molecular docking study was carried out with HEX 6.3 software.Citation26
MTT assay
Normal human peripheral lymphocytes from blood samples were isolated following gradient centrifugation on Ficoll-Paque PLUS as described previously.Citation27
Lymphocytes (1×104 cells/well) were cultured in complete RPMI-1640 media and incubated with different concentrations of CNPs (1, 10, 50, 100, and 200 µg/mL) for 48 h to explore their antiproliferative effect by using MTT assay.Citation27
Flow cytometry analysis
Flow cytometric assay of control and CNP (200 µg/mL)-incubated cells was carried out to quantify the percentage of the apoptotic cells. Cells were stained with fluorescein isothiocyanate (FITC)-Annexin V and propidium iodide (PI) by the flow cytometry kit according to the manufacturer’s instruction (Abcam-Germany). Lymphocytes were incubated with 200 µg/mL CNPs for 48 h at 37°C and 5% CO2. Afterwards, the cells were collected, washed, resuspended in Annexin-binding buffer, and stained with Annexin V-FITC and PI in the dark. Finally, cells were immediately analyzed using a BD FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA).
Statistical analysis
Statistical analysis was undertaken by Student’s t-test. Data were reported as mean ± standard errors (SEs). P-values of <0.05 were reported as significant.
Results and discussion
Characterizations of NPs
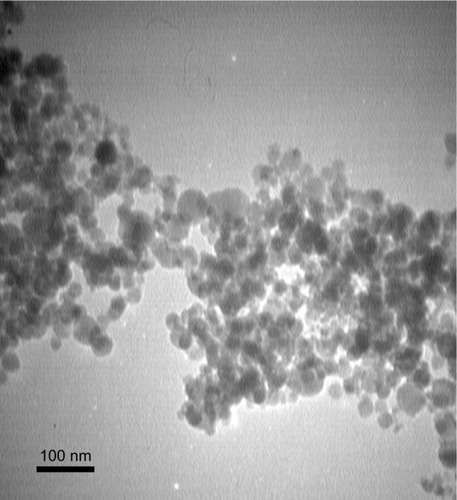
The morphology of the CNP was characterized by the TEM image (). From the TEM image result, it can be observed that a large quantity of uniformed NPs had an average particle size of ~30 nm.
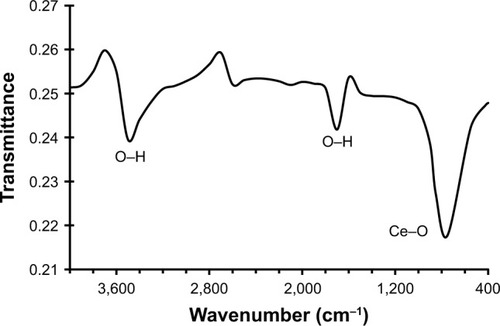
shows the FTIR spectrum of CNP. It shows three strong bands around 3,449, 1,635, and 783 cm−1. The observed bands are similar to those already reported in Masui et al study.Citation28 The vibration bands are at 738 (Ce–O stretching) and 1,635 cm−1 (H2O bending vibration), with a broad peak at 3,478 cm−1 (H2O stretching).
Fluorescence studies
Fluorescence spectroscopy is considered as a promising technique for studying the different aspects of protein-NP interactions. Intrinsic fluorescence intensity of the HHb originated from aromatic residues such as Trp, Tyr and Phe residues. Fluorescence quenching occurs through different possibilities such as static or dynamic mechanisms. Static quenching is defined as the complex formation between the ligand and the protein in the ground state. On the other hand, dynamic quenching occurs when a transient complex between the ligand and protein forms in the excited state. Dynamic or static quenching shows different reactions against temperature and viscosity.Citation29
The Stern–Volmer equation has been widely used for the determination of quenching mechanism of proteins in the presence of ligands.Citation30
Figure 3 Fluorescence titration curve of HHb (2.5 µM) with the various concentrations of CNPs (0, 2.5, 5, 7.5, 10, 12.5, 17.5, 22.5, and 27.5 µM) in phosphate buffer at pH 7.4 and 10 mM at 289 K (A), 310 (B), and 315 K (C).
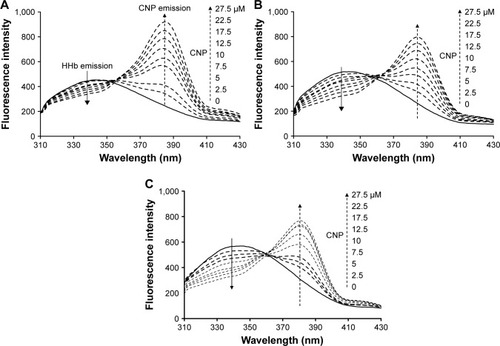
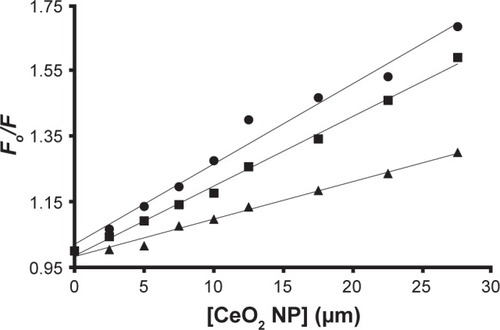
The KSV values for the HHb–CNP complex at 298, 310, and 315 K were 11.4±1.9×103 M−1, 21.2±3.2×103 M−1 and 24.4±3.8×103 M−1, respectively (, ). The direct relation between temperature and KSV indicates the presence of a dynamic quenching mechanism between HHb and CNP. Moreover, dynamic and static quenching can be determined on the basis of the maximum collision-quenching rate constant (kq) of different NPs with the macromolecules, which is 2.0×1010 M−1s−1.Citation21,Citation24
Table 1 KSV and kq values of the fluorescence quenching of HHb (2.5 µM) in the presence of various concentrations of CNPs (0, 2.5, 5, 7.5, 10, 12.5, 17.5, 22.5, and 27.5 µM) at three different temperatures
Figure 4 Stern–Volmer plots for the interaction of CNPs and HHb in phosphate buffer at pH 7.4 and 10 mM at 298 (▲), 310 (■), and 315 K (●).
Based on the following equation:
Binding parameters
The binding parameters for the interaction of CNP with HHb were calculated on the basis of the fluorescence quenching data by using the following Equationequation (3)(3) :Citation31
Table 2 The number of binding sites of protein (n) per NP and Kb of HHb upon interaction with CNPs at three different temperatures of 298, 310, and 315 K
Furthermore, it was reported that n increases from 0.97±0.1 to 1.78±0.1 as the temperature increases from 298 to 315 K. Increasing temperature accelerates the protein unfolding characterized by an exposed hydrophobic core and some loss of quaternary structure. After it passed through the interaction state, the hydrophobic protein core rapidly became more disrupted and the binding-site loop was highly exposed. At higher temperatures, the protein reached a partially denatured state wherein there was virtually some unfolded structure, although fluctuating secondary and native quaternary structures existed. At lower temperatures, a sliding motion of the helix structure did not disrupt the hydrophobic patches of protein, and the interaction binding was weaker at lower temperatures as compared to higher temperatures. To prove that the hydrophobic interaction may play an important role in the interaction between CNPs and HHb, thermodynamic parameters was calculated. Based on the sign of standard enthalpy change (ΔH°) and standard entropy change (ΔS°), the kind of interaction can be determined.Citation32
Thermodynamic parameters
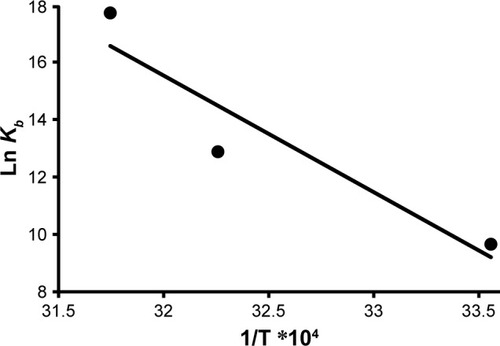
Considering the fact that the enthalpy change (ΔH°) remains almost unchanged over the temperature range, ΔH° and ΔS° can be calculated using the van’t Hoff EquationEquation (4)(4) .Citation33
ΔH° and ΔS° were estimated from the slope and the y-intercept of the plot of ln Kb vs 1/T, respectively ().
Standard free Gibbs energy (ΔG°) was then calculated from the Gibbs–Helmholtz EquationEquation (5)(5) Citation34
The estimated data are summarized in . The negative sign for ΔG° indicated that the binding process between HHb and CNPs was spontaneous. Positive values of ΔH° and ΔS° revealed that hydrophobic forces are involved as the main interaction bonds between CNPs and HHb.Citation35
Table 3 Thermodynamic parameters of HHb upon interaction with CNP at three different temperatures of 298, 310, and 315 K
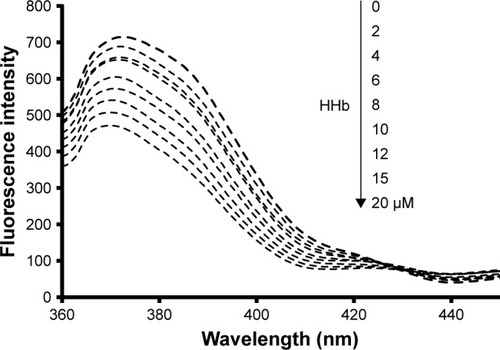
To further support the premise that hydrophobic interaction may play an important role in the formation of the CNP–HHb complex, a fluorescence quenching study was conducted. For this experiment, a fixed concentration of CNPs (5 µM) was used and different concentrations of HHb (2–20 µM) were added to the CNP solution. The excitation wavelength was set to 320 nm. CNPs show strong adsorption intensity at 320 nm (data not shown). As shown in , after the addition of HHb, a quenching of CNP fluorescence intensity was observed. Moreover, a remarkable blue shift (4 nm) was detected as the concentration of HHb increased. This significant blue shift indicates that CNPs have experienced a hydrophobic environment in the presence of HHb, and hydrophobic interaction may play a considerable role in the interaction of CNPs with HHb.
Synchronous fluorescence spectroscopy
Synchronous fluorescence spectroscopy is a useful technique in many biological systems to reveal the microenvironmental changes around aromatic residues. Synchronous fluorescence spectroscopy depends on a multitude of factors such as distance between the donor and acceptor and polarity around residues.Citation35,Citation36 Synchronous fluorescence spectroscopy demonstrates the microenvironmental alterations of Tyr and Trp residues when the Δλ (λem−λex) is 20 and 60 nm, respectively. The effect of CNPs on the synchronous fluorescence spectra of HHb is depicted in . It is indicated that, as the concentration of CNPs increases, the fluorescence intensity of HHb for both Δλ = 20 and 60 nm decreases. However, no remarkable shift in the emission spectrum of HHb was detected. These data suggest that polarity and hydrophobicity around the Trp and Tyr residues remain constant. Briefly, the synchronous fluorescence spectra indicated that the conformation of HHb has not been markedly altered around Tyr and Trp residues.
CD studies
The secondary and quaternary structural changes of HHb upon interaction with CNPs were investigated by far and near UV-CD, respectively.
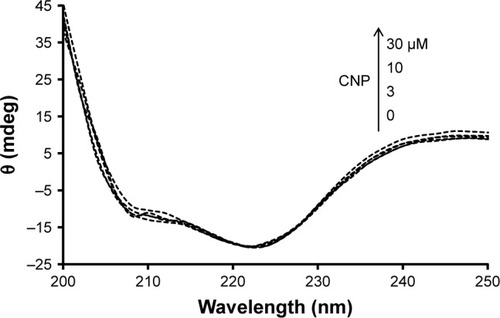
shows the far UV-CD spectra (200–250 nm) of HHb in the absence and presence of different concentrations of CNPs at room temperature. The CD spectrum of HHb shows two characteristic minima at 208 and 222 nm, reflecting the α-helical structure of the HHb.Citation37–Citation39 shows that the secondary structure of HHb has not experienced substantial structural changes after the addition of different concentrations of CNP. This data indicated that the regular secondary structure of the HHb remained unchanged after interaction with CNP. In other words, it may be proposed that the interaction of CNPs and HHb has not led to the NP-induced secondary structural changes of HHb, and the native secondary structure of HHb is preserved.
Figure 8 Far UV-CD spectra (200–250 nm) of HHb (3 µM) were measured in the absence and presence of different CNP concentrations of 3, 10, and 30 µM in phosphate buffer at pH 7.4 and 10 mM at temperature 25°C.
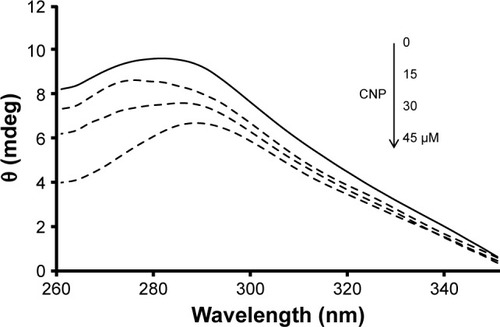
Furthermore, near UV-CD (260–350) was employed to reveal the quaternary structural changes of HHb after interaction with CNP. The position of aromatic residues inside the protein can play considerable roles in the intensity of near UV-CD bands. shows that the band intensities at 285 nm (Trp and Tyr) provide a negligible red shift (285–288 nm) after the addition of CNPs.
Figure 9 Near UV-CD spectra (260–350 nm) of HHb (15 µM) were measured in the absence and presence of different concentrations of CNPs 15, 30, and 45 µM in phosphate buffer at pH 7.4 and 10 mM at temperature 25°C.
Synchronous fluorescence spectroscopy also showed that the polarity around Trp and Tyr was not significantly altered.
As shown in , the quaternary structure of HHb is marginally changed after the addition of CNPs. In other words, Phe, Trp, and Tyr residues have been subjected to a minor displacement inside the protein structure. Therefore, it can be concluded that a large number of Trp, Tyr, and Phe residues in the HHb experience negligible polarity changes. Moreover, it can be deduced that aromatic residues placed close to the surface moved to the polar environment, and this microenvironmental change cannot be detected by CD and fluorescence spectroscopic methods. The displacement of the helix structure of protein and a corresponding movement of aromatic residues to the different polar environment increases the overlap between positive and negative CD bands and finally results in reduced intensity of CD bands.
HHb has 30 Phe (33, 36, 43, 46, 98, 117, 128, 182, 183, 186, 212, 226, 244, 259, 263, 320, 323, 330, 333, 385, 404, 415, 469, 470, 473, 499, 513, 531, 546, and 550), six Trp (14, 156, 178, 301, 443, and 465), and 12 Tyr residues (24, 42, 140, 176, 271, 286, 311, 329, 427, 463, 558, and 573; ).
Figure 10 Plot of relative solvent accessibility area of HHb residues (PDB ID: 2 HHB) from 0 to 1. Color scheme used is: blue, positively charged residues (R, K, and H); red, negatively charged residues (D and E); green, polar uncharged residues (G, N, Y, Q, S, T, and W); yellow, cysteine; gray, hydrophobic residues (all others). This plot was generated by http://www.abren.net/asaview/.
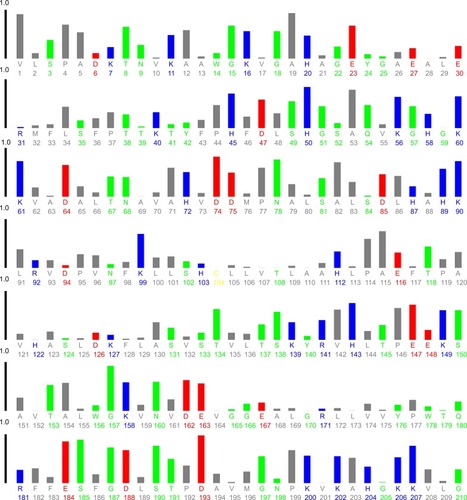
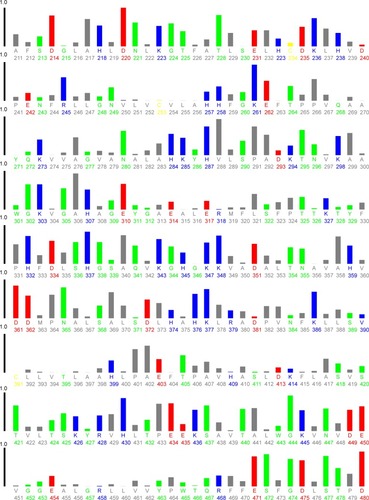
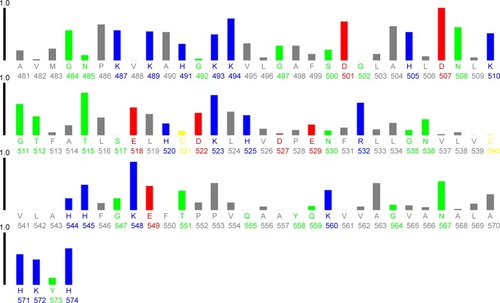
The near UV-CD experiment indicated that the hydrophobicity and polar environment around aromatic residues has not been significantly altered. It may be proposed that Phe residues 182 (β1 subunit) and 469 (β2 subunit), Trp residues 14 (α1 subunit), 156 (β1 subunit), 301 (α2 subunit), and 465 (β2 subunit), and Tyr residues 286 (β1 subunit), 463 (β2 subunit), and 573 (β2 subunit) are more accessible to the solvent and are mainly involved in the interaction of CNPs with HHb ().
To elucidate the accessible sites of HHb for NP interaction, a docking study was conducted.
Docking study
Molecular docking studies have received a great interest in protein–ligand studies. The molecular docking was carried out with both clusters of 1.5 () and 1 nm (), respectively. and show the docking results of CNPs with the size of 1.5 and 1 nm and HHb (PDB ID: 2H35), respectively. The calculated binding energies were found to be −651.85 and −540.90 E-value for larger and smaller CNP clusters, respectively, revealing a greater binding affinity of HHb for larger clusters. Graphical tools such as CHIMERA (www.cgl.ucsf.edu/chimera) and PyMOL (http://pymol.sourceforge.net/) were employed to visualize the docked site. The docked poses for CNPs 1.5 nm/HHb and CNPs 1 nm/HHb are displayed in and , respectively. The CNPs (1.5 nm) with interacting residues (4 Å) are shown in . The closest amino acids to larger CNPs are Lys-139, Thr-134, Thr-137, Lys-127, Pro-95, Ala-130, Phe-98, Ser-102, and Lys-99 in the α-subunit and Trp-37 and Tyr-35 in the β-subunit.
Figure 11 The CNP cluster with the size of 1.5 nm (A); docked site of interaction between CNPs and HHb (B); the CNPs with interacting residues (4 Å) in two rotational views (C and D).
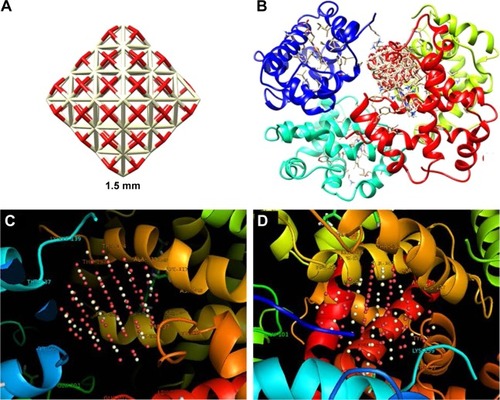
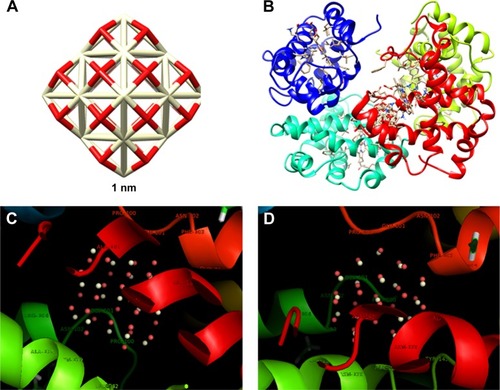
Figure 12 The CNP cluster with the size of 1 nm (A); docked site of interaction between CNPs and HHb (B); the CNPs with interacting residues (4 Å) in two rotational views (C and D).
Tyr-35 and Trp-37 in the β2-subunit can be renumbered as Tyr-463 and Trp-465, which are in good agreement with the plot of accessible surface area and experimental data.
Therefore, it can be seen that some of interacting residues are aromatic amino acids. Moreover, aromatic residues as well as Ala and Pro residues may provide a hydrophobic environment for the interaction of HHb with CNP.
The closest amino acids to smaller CNPs are Pro-100, Glu-101, Asn-102, Phe-103, Ala-142, Asn-133, Ala-141, Ala-138, Asn-139, and Arg-104 (β-subunit; ).
As evident from the results, data obtained from the larger cluster is more oriented toward experimental data, compared with smaller counterparts. This may be due to the fact that NPs tend to form aggregate species in the colloidal state. Therefore, to validate the theoretical studies, larger NPs should be considered for NP–protein interaction.
Furthermore, CNPs can marginally destabilize the quaternary structure of HHb as measured by near UV-CD. However, the secondary structure of protein was not altered as detected by far UV-CD. Generally, the interaction of CNPs induces an overall structural readjustment in the HHb, but keeps the α-helix scaffold intact. This conformational destabilization may cause regulatory dysfunctions of this type of a CNP–HHb complex. The CNP-induced destabilization of HHb suggests that, in the absence of other factors that may influence its stability in vivo such as the protein corona, pH, ionic strength, and opsonization process, the probability of CNP-induced conformational changes of HHb is reduced. Molecular binding of protein onto the NP surface plays a crucial role in the regulation of protein function in the biological systems. NP-adsorbed proteins change their interactions with other ligands at the molecular level. Indeed, the interaction results in minor or substantial alterations in the structure of the protein, compared with their free counterparts, with immediate interference with their function. The formation of a protein–NP system leads to additional and different thermodynamic and conformational parameters; compared to the free forms of protein and NP partners, both processes – NP binding and NP-induced structural changes – are closely correlated. This is more pronounced in the case of multi-subunit proteins with intra- and inter-molecular bonds such that their monomers are denatured, and the system of stability, association, and NP interactions are heavily connected. Thus, NP-binding domains may experience local fluctuation and global unfolding events upon binding to their operator residues.
There were small variations in the displacement of the main aromatic residues as revealed by fluorescence spectroscopy. In addition, a comprehensive analysis by near CD spectra demonstrated minor changes of protein conformation upon interaction with CNPs. These outcomes suggest that the changes in native HHb mediated by CNPs occur on a local basis, perhaps, mostly in the exposed aromatic residues which would not induce a distinct conformational alteration. The interface would stay constant, as indicated by the minor change in the center of maximum wavelength, dramatically inferring no variation in the displacement of the internal aromatic residues to the solvent.
Exploring the secondary structure of HHb by far UV-CD indicated a negligible conformational changes in the presence of CNPs. The molecular basis for this event could be the hydrophobic and hydrophilic interactions of the many residues in the pocket site of the native conformation with the CNP surface. Therefore, the CNPs may bind more specifically through oxygen groups on their surface, somehow mimicking the oxygen molecules in the bloodstream.
It has been reported that functionalized carbon nanotubes induced substantial conformational changes in HHb structure, regardless of single-wall or multi-wall structures.Citation40,Citation41 Moreover, it was shown that nanodiamonds can drive secondary structural changes in HHb structure in a concentration-dependent manner.Citation20 However, this study demonstrated that CNPs do not perturb the secondary structure of Hb.
Nerveless, before we can utilize CNP application in biological and medical systems, cellular assays should be conducted to explore the safety of NPs.
MTT assay
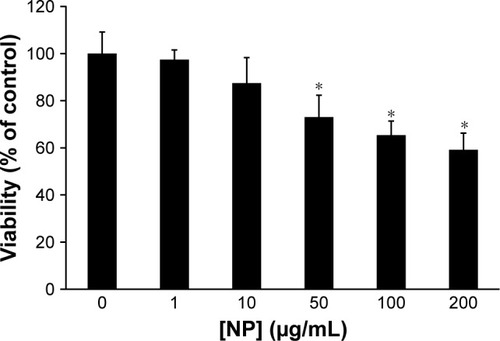
Lymphocytes were used to explore the cytotoxic effect of CNPs. Lymphocytes were incubated with different concentrations of CNPs for 48 h. Cell mortality was evaluated by an MTT assay. It was observed that CNPs suppressed proliferation of lymphocytes (). The cell viability was 100%±9.11%, 97.44%±4.12%, 87.50%±10.76%, 73.06%±9.26%, 65.39%±5.97%, and 59.16%±7.08% after treatment with different concentrations of CNPs from 0 to 1, 10, 50, 100, and 200 µg/mL, respectively. Data displayed in indicated that CNPs (200 µg/mL) induced a remarkable reduction in the cell viability of lymphocytes (*P < 0.05) as compared to control. As demonstrated in , some cytotoxicity was already determined at the 50 µg/mL concentration of CNPs, whereas a maximal effect was revealed at a concentration of 200 µg/mL in lymphocytes. Based on these data, a CNP concentration of 200 µg/mL was used for the flow cytometry test.
Flow cytometry assay
Apoptosis, autophagy, and necrosis are known as the leading mechanisms of cell mortality. To calculate the amount of apoptotic and necrotic cells, the cells were incubated with 200 µg/mL of CNPs for 48 h () and evaluated by flow cytometry. The results were compared with those of the control group (). It was revealed that CNPs can initiate apoptosis in lymphocytes. The flow cytometry outcome demonstrated that the rate of apoptosis increased from 8%±1.33% to 50.67%±7.23% (***P < 0.001) in cells treated with 200 µg/mL of CNPs after 48 h ().
Figure 14 Apoptosis induced by CNPs in lymphocytes. Lymphocytes were incubated with (A) vehicle control and (B) 200 µg/mL CNPs for 48 h. Afterwards, flow cytometric analysis was conducted. (C) Results showing the percentage of viable, apoptotic, and necrotic cells. Data are expressed as mean ± SE (n = 3). *P < 0.05 and ***P < 0.001 vs control group.
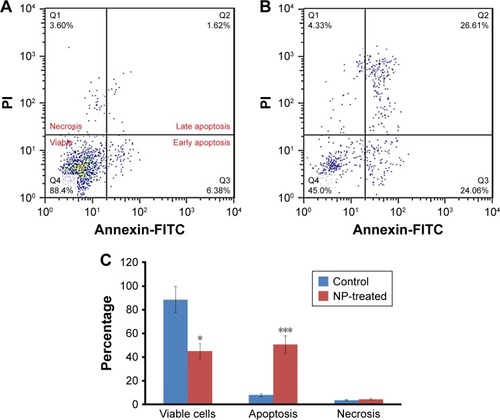
Therefore, it may be indicated that the apoptosis pathway is triggered by CNPs in high concentrations.
Necrosis is initiated by membrane leakage, and apoptosis is mediated through mitochondrial damage. Therefore, it can be suggested that CNPs, after interaction with cells, can mediate cytotoxic effects through mitochondrial pathways.
Consistent with our data, previous reports have indicated that CNPs increase the level of oxidant spices and may trigger apoptosis in human lung epithelial cells.Citation42,Citation43 Therefore, CNPs may initiate cytotoxic impacts upon cellular infiltration. It has been well documented that CNPs of different dimensions markedly exhibited adverse impacts on human hepatoma cells and human neutrophils due to oxidative stress.Citation44,Citation45 Recently, Khan et al also exhibited the cytotoxic effects of CNPs against a colorectal cancer cell line (HT29) through apoptosis induction.Citation46 Forest et al demonstrated that CNPs of various morphologies triggered different mechanisms of cellular toxicity.Citation47
Besides these, CNPs have been revealed to protect cells from oxidative stress due to their potential antioxidant features.Citation48–Citation52 With these conflicting outcomes, the toxicity of CNPs remains difficult to identify, and distinctive adverse effect targets relevant to human safety needs to be explored. Therefore, it is prudent to develop a systematic investigation to explore, in detail, the molecular mechanism underlying the toxicity of CNPs.
Together, these findings exhibited that although CNPs did not perturb the conformation of HHb, they can induce some cellular adverse effects at high concentrations that may limit the medicinal and biological application of CNPs.
Future perspectives
Current uncertainties in nanomaterial assessment in biological systems include NP characterization (size and colloidal stability) and the concentration that correctly delineates and envisages their impacts, and whether they interact with living systems similar to classical xenobiotics. An essential part of investigation quality and standardization with nanomaterials is their characterization. Moreover, an “appropriate” investigation should be conducted in vivo to better explore the fate of NPs upon entry into the body. To better determine the pharmacodynamic and pharmacokinetic parameters of CNPs, a detailed study should be designed to investigate the impact of dimension, morphology, and surface chemistry of CNPs on their distribution and clearance within/from the blood.
For example, how the HHb–CNP interaction can take place in vivo, because the Hb is encapsulated within the red blood cells (RBCs) and their membranes are impermeable to the large NPs. However, it may be difficult to correlate the size of NPs with their ability to cross the RBC membrane, because large NPs may damage the membrane, thereby increasing the membrane permeability.
Furthermore, it is not well explored that how CNPs stimulate apoptosis, necrosis, and autophagy in lymphocytes. NP-induced oxidative stress in mitochondria is dependent on the permeability of the cytoplasmic membrane. Therefore, extrinsic and intrinsic cell death pathways should be investigated to determine the signal cascade between mitochondria and the cell surface.
Conclusion
This research study highlights the reactivity of CNPs with biological systems such as blood components. It was displayed that CNPs affect HHb structure and lymphocyte viability. Herein, the interaction of CNPs with HHb and lymphocytes was studied, and it was reported that CNPs induced marginal structural changes of HHb. Moreover, it was shown that CNPs stimulate cytotoxicity against lymphocytes at high concentrations. The interaction of NPs with proteins and cells can provide useful information about NP bio-reactivity. This interaction results in the formation of a complex NP–protein corona. The protein corona may affect protein structure, such as through secondary structural changes, quaternary structural changes, aggregation, heme degradation, and release of metallic ions. The NP-induced cell mortality may affect the overall bio-reactivity of the NPs. Therefore, in depth investigation of such interactions can play a potential role toward generating bio-compatible NPs with defined and controlled surface characteristics in a biological system. The main aim of this study is to provide important knowledge on the NP–blood component interaction.
Acknowledgments
This research was supported by the Tehran University of Medical Sciences & Health Services (grant no 96033036151), Tehran, Iran.
Disclosure
The authors report no conflicts of interest in this work.
References
- AbdelhameedASAlamPKhanRHBinding of Janus kinase inhibitor tofacitinib with human serum albumin: multi-technique approachJ Biomol Struct Dyn20163492037204426440737
- Radauer-PreimlIAndoschAHawranekTNanoparticle-allergen interactions mediate human allergic responses: protein corona characterization and cellular responsesPart Fibre Toxicol201613131026772182
- IramNKhanMSJollyRInteraction mode of polycarbazole–titanium dioxide nanocomposite with DNA: molecular docking simulation and in-vitro antimicrobial studyJ Photochem Photobiol B2015153203226386641
- NasirZShakirMWahabRCo-precipitation synthesis and characterization of Co doped SnO 2 NPs, HSA interaction via various spectroscopic techniques and their antimicrobial and photocatalytic activitiesInt J Biol Macromol201794Pt A55456527771412
- YeNWangZWangSPeijnenburgWJGMToxicity of mixtures of zinc oxide and graphene oxide nanoparticles to aquatic organisms of different trophic level: particles outperform dissolved ionsNanotoxicology201812542343829658385
- WangZZhangFWangSPeijnenburgWJGMAssessment and prediction of joint algal toxicity of binary mixtures of graphene and ionic liquidsChemosphere201718568168928728125
- WangZWangSPeijnenburgWJGMPrediction of joint algal toxicity of nano-CeO2/nano-TiO2 and florfenicol: independent action surpasses concentration additionChemosphere201615681327156210
- NelsonBCJohnsonMEWalkerMLRileyKRSimsCMAntioxidant cerium oxide nanoparticles in biology and medicineAntioxidants (Basel)201652 pii:E15
- BaileyZSNilsonEBatesJACerium oxide nanoparticles improve outcome after in vitro and in vivo mild traumatic brain injuryJ Neurotrauma Epub2016112
- ZhouDFangTLuLQYiLNeuroprotective potential of cerium oxide nanoparticles for focal cerebral ischemic strokeJ Huazhong Univ Sci Technolog Med Sci201636448048627465320
- PopovALPopovaNRSeleznevaIIAkkizovAYIvanovVKCerium oxide nanoparticles stimulate proliferation of primary mouse embryonic fibroblasts in vitroMater Sci Eng C Mater Biol Appl20166840641327524035
- AzariAShokrzadehMZamaniEAmaniNShakiFCerium oxide nanoparticles protects against acrylamide induced toxicity in HepG2 cells through modulation of oxidative stressDrug and Chemical Toxicology201816
- PezziniIMarinoADel TurcoSCerium oxide nanoparticles: the regenerative redox machine in bioenergetic imbalanceNanomedicine (Lond)201712440341628000542
- NaganumaTShape design of cerium oxide nanoparticles for enhancement of enzyme mimetic activity in therapeutic applicationsNano Res2017101199217
- KhaksarMRRahimifardMBaeeriMProtective effects of cerium oxide and yttrium oxide nanoparticles on reduction of oxidative stress induced by sub-acute exposure to diazinon in the rat pancreasJ Trace Elem Med Biol201741799028347467
- GilDRodriguezJWardBVertegelAIvanovVReukovVAntioxidant activity of SOD and catalase conjugated with nanocrystalline ceriaBioengineering (Basel)2017411825
- ManningLRPopowiczAMPadovanJCChaitBTManningJMGel filtration of dilute human embryonic hemoglobins reveals basis for their increased oxygen bindingAnal Biochem2017519384127965062
- TomaVAFarcaşADRomanIComparative in vivo effects of hemoglobin-based oxygen carriers (HBOC) with varying prooxidant and physiological reactivityPLoS One2016114e015390927097326
- MauryaNMauryaJKKumariMKhanABDohareRPatelRHydrogen bonding-assisted interaction between amitriptyline hydrochloride and hemoglobin: spectroscopic and molecular dynamics studiesJ Biomol Struct Dyn20173561367138027141981
- PishkarLTaheriSMakaremSStudies on the interaction between nanodiamond and human hemoglobin by surface tension measurement and spectroscopy methodsJ Biomol Struct Dyn201735360361527151742
- Jafari AzadVKasraviSAlizadeh ZeinabadHProbing the conformational changes and peroxidase-like activity of cytochrome c upon interaction with iron nanoparticlesJ Biomol Struct Dyn201735122565257727632558
- ZeinabadHAZarrabianASabouryAAAlizadehAMFalahatiMInteraction of single and multi wall carbon nanotubes with the biological systems: tau protein and PC12 cells as targetsSci Rep201662650827216374
- AghiliZTaheriSZeinabadHAInvestigating the interaction of Fe nanoparticles with lysozyme by biophysical and molecular docking studiesPLoS One20161110e016487827776180
- ZeinabadHAKachooeiESabouryAAThermodynamic and conformational changes of protein toward interaction with nanoparticles: a spectroscopic overviewRSC Adv20166107105903105919
- LoschenCMiganiABromleySTIllasFNeymanKMDensity functional studies of model cerium oxide nanoparticlesPhys Chem Chem Phys200810375730573818956108
- RitchieDWVenkatramanVUltra-fast FFT protein docking on graphics processorsBioinformatics201026192398240520685958
- MoosaviMASharifiMGhafarySMPhotodynamic N-TiO 2 nanoparticle treatment induces controlled Ros-mediated autophagy and terminal differentiation of leukemia cellsSci Rep201663441327698385
- MasuiTHiraiHHamadaRSynthesis and characterization of cerium oxide nanoparticles coated with turbostratic boron nitrideJ Mater Chem2003133622627
- CarusoÍPBarbosa FilhoJMde AraújoASde SouzaFPFosseyMACornélioMLAn integrated approach with experimental and computational tools outlining the cooperative binding between 2-phenylchromone and human serum albuminFood Chem201619693594226593575
- WuZShenLHanQMechanism and nature of inhibition of trypsin by ligupurpuroside A, a Ku-Ding tea extract, studied by spectroscopic and docking methodsFood Biophys20171217887
- RabbaniGBaigMHJanATBinding of erucic acid with human serum albumin using a spectroscopic and molecular docking studyInt J Biol Macromol2017105Pt 31572158028414112
- Rahimi-VagharRDivsalarAMansouri-TorshiziHHeme releasing from human hemoglobin upon interaction with a new synthesized complex of 1, 10-phenanthroline-n-butyl dithiocarbamato pd (ii) nitratePhys Chem Res201312185196
- ChenWYHuangHMLinCCLinFYChanYCEffect of temperature on hydrophobic interaction between proteins and hydrophobic adsorbents: studies by isothermal titration calorimetry and the van’t Hoff equationLangmuir2003192293959403
- PerozzoRFolkersGScapozzaLThermodynamics of protein–ligand interactions: history, presence, and future aspectsJ Recept Signal Transduct Res2004241–215215344878
- AjmalMRAbdelhameedASAlamPKhanRHInteraction of new kinase inhibitors cabozantinib and tofacitinib with human serum alpha-1 acid glycoprotein. A comprehensive spectroscopic and molecular Docking approachSpectrochim Acta A Mol Biomol Spectrosc201615919920826851488
- SabziparvarNSaeediYNouriMInvestigating the Interaction of Silicon Dioxide Nanoparticles with Human Hemoglobin and Lymphocyte Cells by Biophysical, Computational, and Cellular StudiesThe Journal of Physical Chemistry B2018122154278428829537841
- BasakPDebnathTBanerjeeRBhattacharyyaMSelective binding of divalent cations toward heme proteinsFront Biol20161113242
- MaheshwariNKhanFHMahmoodRSodium meta-arsenite induced reactive oxygen species in human red blood cells: impaired antioxidant and membrane redox systems, haemoglobin oxidation and morphological changesFree Radic Res201751548349728480809
- ChatterjeeSKumarGSBinding of fluorescent acridine dyes acridine orange and 9-aminoacridine to hemoglobin: elucidation of their molecular recognition by spectroscopy, calorimetry and molecular modeling techniquesJ Photochem Photobiol B201615916917827077554
- WangYQZhangHMCaoJBinding of hydroxylated single-walled carbon nanotubes to two hemoproteins, hemoglobin and myoglobinJ Photochem Photobiol B2014141263525313539
- SekarGKandiyilSTSivakumarAMukherjeeAChandrasekaranNBinding studies of hydroxylated Multi-Walled Carbon Nanotubes to hemoglobin, gamma globulin and transferrinJ Photochem Photobiol B201515322223226432959
- LinWHuangYWZhouXDMaYToxicity of cerium oxide nanoparticles in human lung cancer cellsInt J Toxicol200625645145717132603
- ParkEJChoiJParkYKParkKOxidative stress induced by cerium oxide nanoparticles in cultured BEAS-2B cellsToxicology20082451–29010018243471
- ChengGGuoWHanLCerium oxide nanoparticles induce cytotoxicity in human hepatoma SMMC-7721 cells via oxidative stress and the activation of MAPK signaling pathwaysToxicol In Vitro20132731082108823416263
- BabinKAntoineFGoncalvesDMGirardDTiO2, CeO2 and ZnO nanoparticles and modulation of the degranulation process in human neutrophilsToxicol Lett20132211576323726862
- KhanSAnsariAARolfoCEvaluation of in vitro cytotoxicity, biocompatibility, and changes in the expression of apoptosis regulatory proteins induced by cerium oxide nanocrystalsSci Technol Adv Mater201718136437328634498
- ForestVLeclercLHochepiedJFTrouvéASarryGPourchezJImpact of cerium oxide nanoparticles shape on their in vitro cellular toxicityToxicol In Vitro20173813614127693598
- SorenSJenaSRSamantaLParhiPAntioxidant potential and toxicity study of the cerium oxide nanoparticles synthesized by microwave-mediated synthesisAppl Biochem Biotechnol2015177114816126137877
- ErikssonPTalAASkallbergACerium oxide nanoparticles with antioxidant capabilities and gadolinium integration for MRI contrast enhancementScientific Reports201881699929725117
- RubioLAnnangiBVilaLHernándezAMarcosRAntioxidant and anti-genotoxic properties of cerium oxide nanoparticles in a pulmonary-like cell systemArch Toxicol201690226927825618551
- XueYBalmuriSRPatelASantVSantSSynthesis, physicochemical characterization, and antioxidant effect of PEGylated cerium oxide nanoparticlesDrug Deliv Transl Res20188235736728589454
- SadhuAGhoshIMoriyasuYMukherjeeABandyopadhyayMRole of cerium oxide nanoparticle-induced autophagy as a safeguard to exogenous H2O2-mediated DNA damage in tobacco BY-2 cellsMutagenesis201833216117729506140